SARS-CoV-2 recombinant proteins stimulate distinct cellular and humoral immune response profiles in samples from COVID-19 convalescent patients
- PMID: 34878034
- PMCID: PMC8610223
- DOI: 10.6061/clinics/2021/e3548
SARS-CoV-2 recombinant proteins stimulate distinct cellular and humoral immune response profiles in samples from COVID-19 convalescent patients
Abstract
Objectives: In this preliminary study we investigated cellular and humoral immune responses to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antigens in blood samples from 14 recovered coronavirus disease 2019 (COVID-19) patients and compared them to those in samples from 12 uninfected/unvaccinated volunteers.
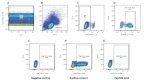
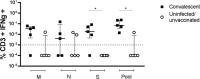
Methods: Cellular immunity was assessed by intracellular detection of IFN-γ in CD3+ T lymphocytes after stimulation with SARS-CoV-2 spike (S1), nucleocapsid (NC), or receptor-binding domain (RBD) recombinant proteins or overlapping peptide pools covering the sequence of SARS-CoV-2 spike, membrane and nucleocapsid regions. The humoral response was examined by ELISAs and/or chemiluminescence assays for the presence of serum IgG antibodies directed to SARS-CoV-2 proteins.
Results: We observed differences between humoral and cellular immune profiles in response to stimulation with the same proteins. Assays of IgG antibodies directed to SARS-CoV-2 NC, RBD and S1/S2 recombinant proteins were able to differentiate convalescent from uninfected/unvaccinated groups. Cellular immune responses to SARS-CoV-2 protein stimuli did not exhibit a specific response, as T cells from both individuals with no history of contact with SARS-CoV-2 and from recovered donors were able to produce IFN-γ.
Conclusions: Determination of the cellular immune response to stimulation with a pool of SARS-CoV-2 peptides but not with SARS-CoV-2 proteins is able to distinguish convalescent individuals from unexposed individuals. Regarding the humoral immune response, the screening for serum IgG antibodies directed to SARS-CoV-2 proteins has been shown to be specific for the response of recovered individuals.
Conflict of interest statement
No potential conflict of interest was reported.
Figures




References
-
- Rabaan AA, Al-Ahmed SH, Haque S, Sah R, Tiwari R, Malik YS, et al. SARS-CoV-2, SARS-CoV, and MERS-COV: A comparative overview. Infez Med. 2020;28(2):174–84. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

