Role of miR-181c in Diet-induced obesity through regulation of lipid synthesis in liver
- PMID: 34879063
- PMCID: PMC8654194
- DOI: 10.1371/journal.pone.0256973
Role of miR-181c in Diet-induced obesity through regulation of lipid synthesis in liver
Abstract
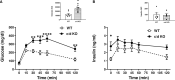
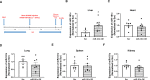
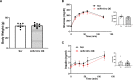
We recently identified a nuclear-encoded miRNA (miR-181c) in cardiomyocytes that can translocate into mitochondria to regulate mitochondrial gene mt-COX1 and influence obesity-induced cardiac dysfunction through the mitochondrial pathway. Because liver plays a pivotal role during obesity, we hypothesized that miR-181c might contribute to the pathophysiological complications associated with obesity. Therefore, we used miR-181c/d-/- mice to study the role of miR-181c in hepatocyte lipogenesis during diet-induced obesity. The mice were fed a high-fat (HF) diet for 26 weeks, during which indirect calorimetric measurements were made. Quantitative PCR (qPCR) was used to examine the expression of genes involved in lipid synthesis. We found that miR-181c/d-/- mice were not protected against all metabolic consequences of HF exposure. After 26 weeks, the miR-181c/d-/- mice had a significantly higher body fat percentage than did wild-type (WT) mice. Glucose tolerance tests showed hyperinsulinemia and hyperglycemia, indicative of insulin insensitivity in the miR-181c/d-/- mice. miR-181c/d-/- mice fed the HF diet had higher serum and liver triglyceride levels than did WT mice fed the same diet. qPCR data showed that several genes regulated by isocitrate dehydrogenase 1 (IDH1) were more upregulated in miR-181c/d-/- liver than in WT liver. Furthermore, miR-181c delivered in vivo via adeno-associated virus attenuated the lipogenesis by downregulating these same lipid synthesis genes in the liver. In hepatocytes, miR-181c regulates lipid biosynthesis by targeting IDH1. Taken together, the data indicate that overexpression of miR-181c can be beneficial for various lipid metabolism disorders.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures








References
-
- Alberti KG, Eckel RH, Grundy SM, Zimmet PZ, Cleeman JI, Donato KA, et al. Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation. 2009;120(16):1640–5. Epub 2009/10/07. doi: 10.1161/CIRCULATIONAHA.109.192644 . - DOI - PubMed
-
- Das S, Bedja D, Campbell N, Dunkerly B, Chenna V, Maitra A, et al. miR-181c regulates the mitochondrial genome, bioenergetics, and propensity for heart failure in vivo. PLoS One. 2014;9(5):e96820. Epub 2014/05/09. doi: 10.1371/journal.pone.0096820 ; PubMed Central PMCID: PMC4014556. - DOI - PMC - PubMed
-
- Das S, Kohr M, Dunkerly-Eyring B, Lee DI, Bedja D, Kent OA, et al. Divergent Effects of miR-181 Family Members on Myocardial Function Through Protective Cytosolic and Detrimental Mitochondrial microRNA Targets. J Am Heart Assoc. 2017;6(3). Epub 2017/03/01. doi: 10.1161/JAHA.116.004694 ; PubMed Central PMCID: PMC5524005. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

