A transcriptional rheostat couples past activity to future sensory responses
- PMID: 34879231
- PMCID: PMC8758202
- DOI: 10.1016/j.cell.2021.11.022
A transcriptional rheostat couples past activity to future sensory responses
Abstract
Animals traversing different environments encounter both stable background stimuli and novel cues, which are thought to be detected by primary sensory neurons and then distinguished by downstream brain circuits. Here, we show that each of the ∼1,000 olfactory sensory neuron (OSN) subtypes in the mouse harbors a distinct transcriptome whose content is precisely determined by interactions between its odorant receptor and the environment. This transcriptional variation is systematically organized to support sensory adaptation: expression levels of more than 70 genes relevant to transforming odors into spikes continuously vary across OSN subtypes, dynamically adjust to new environments over hours, and accurately predict acute OSN-specific odor responses. The sensory periphery therefore separates salient signals from predictable background via a transcriptional rheostat whose moment-to-moment state reflects the past and constrains the future; these findings suggest a general model in which structured transcriptional variation within a cell type reflects individual experience.
Keywords: Act-seq; adaptation; functional imaging; gene expression programs; homeostasis; odor coding; odorant receptor; olfaction; sensory neurons; single-cell RNA sequencing; transcription.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

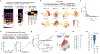


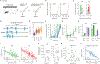


References
-
- Attneave F (1954). Some informational aspects of visual perception. Psychol Rev 61, 183–193. - PubMed
-
- Barlow H (1961). Possible principles underlying the transformation of sensory messages. In Sensory Communication (Cambridge, MA: MIT press; ), pp. 217–234.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

