Exercise plasma boosts memory and dampens brain inflammation via clusterin
- PMID: 34880498
- PMCID: PMC9721468
- DOI: 10.1038/s41586-021-04183-x
Exercise plasma boosts memory and dampens brain inflammation via clusterin
Abstract
Physical exercise is generally beneficial to all aspects of human and animal health, slowing cognitive ageing and neurodegeneration1. The cognitive benefits of physical exercise are tied to an increased plasticity and reduced inflammation within the hippocampus2-4, yet little is known about the factors and mechanisms that mediate these effects. Here we show that 'runner plasma', collected from voluntarily running mice and infused into sedentary mice, reduces baseline neuroinflammatory gene expression and experimentally induced brain inflammation. Plasma proteomic analysis revealed a concerted increase in complement cascade inhibitors including clusterin (CLU). Intravenously injected CLU binds to brain endothelial cells and reduces neuroinflammatory gene expression in a mouse model of acute brain inflammation and a mouse model of Alzheimer's disease. Patients with cognitive impairment who participated in structured exercise for 6 months had higher plasma levels of CLU. These findings demonstrate the existence of anti-inflammatory exercise factors that are transferrable, target the cerebrovasculature and benefit the brain, and are present in humans who engage in exercise.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Figures


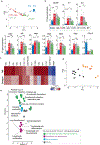

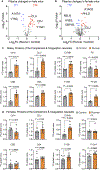


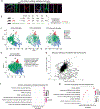


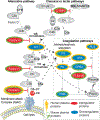



Comment in
-
Exercise-induced plasma factors confer brain benefits.Nat Rev Endocrinol. 2022 Feb;18(2):68. doi: 10.1038/s41574-021-00624-9. Nat Rev Endocrinol. 2022. PMID: 34931019 No abstract available.
-
An exercise infusion benefits brain function.Cell Res. 2022 Mar;32(3):223-224. doi: 10.1038/s41422-022-00621-1. Cell Res. 2022. PMID: 35075253 Free PMC article. No abstract available.
References
-
- Prakash RS, Voss MW, Erickson KI & Kramer AF Physical activity and cognitive vitality. Annu. Rev. Psychol 66, 769–797 (2015). - PubMed
-
- Gleeson M et al. The anti-inflammatory effects of exercise: mechanisms and implications for the prevention and treatment of disease. Nat. Rev. Immunol 11, 607–615 (2011). - PubMed
-
- Hawley JA, Hargreaves M, Joyner MJ & Zierath JR Integrative biology of exercise. Cell 159, 738–749 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

