Conservation and Divergence of the CONSTANS-Like (COL) Genes Related to Flowering and Circadian Rhythm in Brassica napus
- PMID: 34880887
- PMCID: PMC8645894
- DOI: 10.3389/fpls.2021.760379
Conservation and Divergence of the CONSTANS-Like (COL) Genes Related to Flowering and Circadian Rhythm in Brassica napus
Abstract
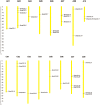
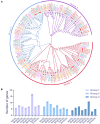
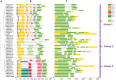
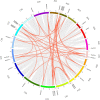
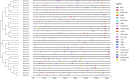
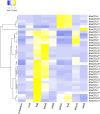
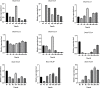
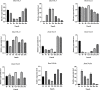
The CONSTANS-LIKE (COL) genes are important signaling component in the photoperiod pathway and flowering regulation pathway. However, people still know little about their role in Brassica napus. To achieve a better understanding of the members of the BnaCOL gene family, reveal their evolutionary relationship and related functions involved in photoperiod regulation, we systematically analyzed the BnaCOL family members in B. napus genome. A total of 33 BnaCOL genes distributed unevenly on 16 chromosomes were identified in B. napus and could be classified into three subfamilies. The same subfamilies have relatively conservative gene structures, three-dimensional protein structures and promoter motifs such as light-responsive cis-elements. The collinearity analysis detected 37 pairs of repetitive genes in B. napus genome. A 67.7% of the BnaCOL genes were lost after B. napus genome polyploidization. In addition, the BnaCOL genes showed different tissue-specific expression patterns. A 81.8% of the BnaCOL genes were mainly expressed in leaves, indicating that they may play a conservative role in leaves. Subsequently, we tested the circadian expression profiles of nine homologous genes that regulate flowering in Arabidopsis. Most BnaCOL genes exhibit several types of circadian rhythms, indicating that these BnaCOL genes are involved in the photoperiod pathway. As such, our research has laid the foundation for understanding the exact role of the BnaCOL family in the growth and development of rapeseed, especially in flowering.
Keywords: Brassica napus; CONSTANS-LIKE (COL) genes; expression pattern; genome-wide analysis; photoperiod.
Copyright © 2021 Chen, Zhou, Hu, Wei and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Genome-wide analysis of maize CONSTANS-LIKE gene family and expression profiling under light/dark and abscisic acid treatment.Gene. 2018 Oct 5;673:1-11. doi: 10.1016/j.gene.2018.06.032. Epub 2018 Jun 13. Gene. 2018. PMID: 29908279
-
Identification and characterization of the CONSTANS (CO)/CONSTANS-like (COL) genes related to photoperiodic signaling and flowering in tomato.Plant Sci. 2020 Dec;301:110653. doi: 10.1016/j.plantsci.2020.110653. Epub 2020 Sep 1. Plant Sci. 2020. PMID: 33218623
-
Genome-wide association study reveals the genetic architecture of flowering time in rapeseed (Brassica napus L.).DNA Res. 2016 Feb;23(1):43-52. doi: 10.1093/dnares/dsv035. Epub 2015 Dec 10. DNA Res. 2016. PMID: 26659471 Free PMC article.
-
Genome-wide identification, expression, and sequence analysis of CONSTANS-like gene family in cannabis reveals a potential role in plant flowering time regulation.BMC Plant Biol. 2021 Mar 17;21(1):142. doi: 10.1186/s12870-021-02913-x. BMC Plant Biol. 2021. PMID: 33731002 Free PMC article.
-
Phylogenomic analysis of 20S proteasome gene family reveals stress-responsive patterns in rapeseed (Brassica napus L.).Front Plant Sci. 2022 Oct 31;13:1037206. doi: 10.3389/fpls.2022.1037206. eCollection 2022. Front Plant Sci. 2022. PMID: 36388569 Free PMC article.
Cited by
-
Genome-Wide Identification of CONSTANS-like (COL) Gene Family and the Potential Function of ApCOL08 Under Salt Stress in Andrographis paniculata.Int J Mol Sci. 2025 Jan 16;26(2):724. doi: 10.3390/ijms26020724. Int J Mol Sci. 2025. PMID: 39859441 Free PMC article.
-
Their C-termini divide Brassica rapa FT-like proteins into FD-interacting and FD-independent proteins that have different effects on the floral transition.Front Plant Sci. 2023 Jan 12;13:1091563. doi: 10.3389/fpls.2022.1091563. eCollection 2022. Front Plant Sci. 2023. PMID: 36714709 Free PMC article.
-
Genome-Wide Identification and Characterization of the CCT Gene Family in Rapeseed (Brassica napus L.).Int J Mol Sci. 2024 May 13;25(10):5301. doi: 10.3390/ijms25105301. Int J Mol Sci. 2024. PMID: 38791340 Free PMC article.
-
Transcriptomic and metabolomic profiling reveals molecular regulatory network involved in flower development and phenotypic changes in two Lonicera macranthoides varieties.3 Biotech. 2024 Jul;14(7):174. doi: 10.1007/s13205-024-04019-1. Epub 2024 Jun 5. 3 Biotech. 2024. PMID: 38855147 Free PMC article.
-
Characterization of Phytohormones and Transcriptomic Profiling of the Female and Male Inflorescence Development in Manchurian Walnut (Juglans mandshurica Maxim.).Int J Mol Sci. 2022 May 13;23(10):5433. doi: 10.3390/ijms23105433. Int J Mol Sci. 2022. PMID: 35628244 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

