Identification of the key functional genes in salt-stress tolerance of Cyanobacterium Phormidium tenue using in silico analysis
- PMID: 34881166
- PMCID: PMC8602552
- DOI: 10.1007/s13205-021-03050-w
Identification of the key functional genes in salt-stress tolerance of Cyanobacterium Phormidium tenue using in silico analysis
Abstract
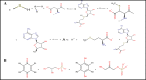
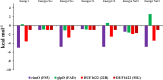
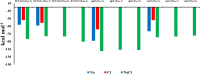
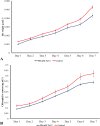
The development of artificial biocrust using cyanobacterium Phormidium tenue has been suggested as an effective strategy to prevent soil degradation. Here, a combination of in silico approaches with growth rate, photosynthetic pigment, morphology, and transcript analysis was used to identify specific genes and their protein products in response to 500 mM NaCl in P. tenue. The results show that 500 mM NaCl induces the expression of genes encoding glycerol-3-phosphate dehydrogenase (glpD) as a Flavoprotein, ribosomal protein S12 methylthiotransferase (rimO), and a hypothetical protein (sll0939). The constructed co-expression network revealed a group of abiotic stress-responsive genes. Using the Basic Local Alignment Search Tool (BLAST), the homologous proteins of rimO, glpD, and sll0939 were identified in the P. tenue genome. Encoded proteins of glpD, rimO, and DUF1622 genes, respectively, contain (DAO and DAO C), (UPF0004, Radical SAM and TRAM 2), and (DUF1622) domains. The predicted ligand included 22B and MG for DUF1622, FS5 for rimO, and FAD for glpD protein. There was no direct disruption in ligand-binding sites of these proteins by Na+, Cl-, or NaCl. The growth rate, photosynthetic pigment, and morphology of P. tenue were investigated, and the result showed an acceptable tolerance rate of this microorganism under salt stress. The quantitative real-time polymerase chain reaction (qRT-PCR) results revealed the up-regulation of glpD, rimO, and DUF1622 genes under salt stress. This is the first report on computational and experimental analyses of the glpD, rimO, and DUF1622 genes in P. tenue under salt stress to the best of our knowledge.
Supplementary information: The online version contains supplementary material available at 10.1007/s13205-021-03050-w.
Keywords: Biocrust; Homology modeling; Molecular docking; NaCl; Phormidium tenue; Transcript analysis.
© King Abdulaziz City for Science and Technology 2021.
Figures


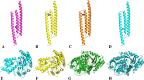



References
-
- Allakhverdiev SI, Murata N. Salt stress inhibits photosystems II and I in cyanobacteria. Photosynth Res. 2008;98:529–539. - PubMed
-
- Antoninka A, Bowker MA, Reed SC, Doherty K. Production of greenhouse-grown biocrust mosses and associated cyanobacteria to rehabilitate dryland soil function. Restor Ecol. 2016;24:324–335.
LinkOut - more resources
Full Text Sources
Research Materials

