Clinical characterization and immunosuppressive regulation of CD161 (KLRB1) in glioma through 916 samples
- PMID: 34881489
- PMCID: PMC8819299
- DOI: 10.1111/cas.15236
Clinical characterization and immunosuppressive regulation of CD161 (KLRB1) in glioma through 916 samples
Abstract
Background: Glioblastoma is a paradigm of cancer-associated immunosuppression, limiting the effects of immunotherapeutic strategies. Thus, identifying the molecular mechanisms underlying immune surveillance evasion is critical. Recently, the preferential expression of inhibitory natural killer (NK) cell receptor CD161 on glioma-infiltrating cytotoxic T cells was identified. Focusing on the molecularly annotated, large-scale clinical samples from different ethnic origins, the data presented here provide evidence of this immune modulator's essential roles in brain tumor biology.
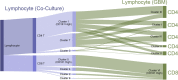
Methods: Retrospective RNA-seq data analysis was conducted in a cohort of 313 patients with glioma in the Chinese Glioma Genome Atlas (CGGA) database and 603 patients in The Cancer Genome Atlas (TCGA) database. In addition, single-cell sequencing data from seven surgical specimens of glioblastoma patients and a model in which patient-derived glioma stem cells were cocultured with peripheral lymphocytes, were used to analyze the molecular evolution process during gliomagenesis.
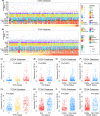
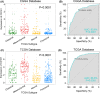
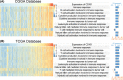
Results: CD161 was enriched in high-grade gliomas and isocitrate dehydrogenase (IDH)-wildtype glioma. CD161 acted as a potential biomarker for the mesenchymal subtype of glioma and an independent prognostic factor for the overall survival (OS) of patients with glioma. In addition, CD161 played an essential role in inhibiting the cytotoxicity of T cells in glioma patients. During the process of gliomagenesis, the expression of CD161 on different lymphocytes dynamically evolved.
Conclusion: The expression of CD161 was closely related to the pathology and molecular pathology of glioma. Meanwhile, CD161 promoted the progression and evolution of gliomas through its unique effect on T cell dysfunction. Thus, CD161 is a promising novel target for immunotherapeutic strategies in glioma treatment.
Keywords: CD161; T cell evolution; glioma; immunotherapy; neuro-immunity; prognosis.
© 2021 The Authors. Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Figures





References
-
- Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209‐249. - PubMed
-
- Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803‐820. - PubMed
-
- Jiang T, Nam D‐H, Ram Z, et al. Clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett. 2021;499:60‐72. - PubMed
-
- Wang H, Xu T, Huang Q, et al. Immunotherapy for malignant glioma: current status and future directions. Trends Pharmacol Sci. 2020;41(2):123‐138. - PubMed
MeSH terms
Substances
Grants and funding
- M-0020/Sino-German Center Cooperation and Exchanges Program
- PXM2019_026280_000002/Construction of the Genomics Platform for Chinese People's Brain Diseases
- 82002994/National Natural Science Foundation of China
- 82072768/National Natural Science Foundation of China
- 2020-I2M-C&T-A-024/The Medical and Health Technology Innovation Project of the Chinese Academy of Medical Sciences
LinkOut - more resources
Full Text Sources
Other Literature Sources

