Inhibition of the PLK1-Coupled Cell Cycle Machinery Overcomes Resistance to Oxaliplatin in Colorectal Cancer
- PMID: 34881526
- PMCID: PMC8655181
- DOI: 10.1002/advs.202100759
Inhibition of the PLK1-Coupled Cell Cycle Machinery Overcomes Resistance to Oxaliplatin in Colorectal Cancer
Abstract
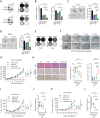
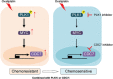
Dysregulation of the cell cycle machinery leads to genomic instability and is a hallmark of cancer associated with chemoresistance and poor prognosis in colorectal cancer (CRC). Identifying and targeting aberrant cell cycle machinery is expected to improve current therapies for CRC patients. Here,upregulated polo-like kinase 1 (PLK1) signaling, accompanied by deregulation of cell cycle-related pathways in CRC is identified. It is shown that aberrant PLK1 signaling correlates with recurrence and poor prognosis in CRC patients. Genetic and pharmacological blockade of PLK1 significantly increases the sensitivity to oxaliplatin in vitro and in vivo. Mechanistically, transcriptomic profiling analysis reveals that cell cycle-related pathways are activated by oxaliplatin treatment but suppressed by a PLK1 inhibitor. Cell division cycle 7 (CDC7) is further identified as a critical downstream effector of PLK1 signaling, which is transactivated via the PLK1-MYC axis. Increased CDC7 expression is also found to be positively correlated with aberrant PLK1 signaling in CRC and is associated with poor prognosis. Moreover, a CDC7 inhibitor synergistically enhances the anti-tumor effect of oxaliplatin in CRC models, demonstrating the potential utility of targeting the PLK1-MYC-CDC7 axis in the treatment of oxaliplatin-based chemotherapy.
Keywords: MYC; cell division cycle 7; colorectal cancer; oxaliplatin; polo-like kinase 1.
© 2021 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
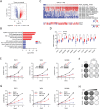
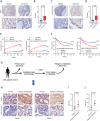
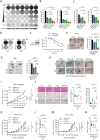
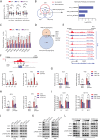

References
Publication types
MeSH terms
Substances
Grants and funding
- 2017YFC1308800/National Key R&D Program of China
- National Key Clinical Discipline
- 81972212/National Natural Science Foundation of China
- 82003197/National Natural Science Foundation of China
- 81972596/National Natural Science Foundation of China
- 81772963/National Natural Science Foundation of China
- 17ykpy66/Fundamental Research Funds for the Central Universities
- 2017A030310517/Guangdong Natural Science Foundation
- 2019A1515010063/Guangdong Natural Science Foundation
- 2018A030310268/Guangdong Natural Science Foundation
- 201803040019/Science and Technology Planning Project of Guangzhou City
- A2018274/Medical Science and Technology Foundation of Guangdong Province
- P20200217202309878/Research Fund of the Sixth Affiliated Hospital of Sun Yat-sen University
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
