Genome-wide host methylation profiling of anal and cervical carcinoma
- PMID: 34882728
- PMCID: PMC8659695
- DOI: 10.1371/journal.pone.0260857
Genome-wide host methylation profiling of anal and cervical carcinoma
Abstract
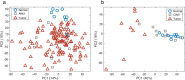
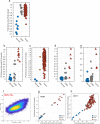
HPV infection results in changes in host gene methylation which, in turn, are thought to contribute to the neoplastic progression of HPV-associated cancers. The objective of this study was to identify joint and disease-specific genome-wide methylation changes in anal and cervical cancer as well as changes in high-grade pre-neoplastic lesions. Formalin-fixed paraffin-embedded (FFPE) anal tissues (n = 143; 99% HPV+) and fresh frozen cervical tissues (n = 28; 100% HPV+) underwent microdissection, DNA extraction, HPV genotyping, bisulfite modification, DNA restoration (FFPE) and analysis by the Illumina HumanMethylation450 Array. Differentially methylated regions (DMR; t test q<0.01, 3 consecutive significant CpG probes and mean Δβ methylation value>0.3) were compared between normal and cancer specimens in partial least squares (PLS) models and then used to classify anal or cervical intraepithelial neoplasia-3 (AIN3/CIN3). In AC, an 84-gene PLS signature (355 significant probes) differentiated normal anal mucosa (NM; n = 9) from AC (n = 121) while a 36-gene PLS signature (173 significant probes) differentiated normal cervical epithelium (n = 10) from CC (n = 9). The CC progression signature was validated using three independent publicly available datasets (n = 424 cases). The AC and CC progression PLS signatures were interchangeable in segregating normal, AIN3/CIN3 and AC and CC and were found to include 17 common overlapping hypermethylated genes. Moreover, these signatures segregated AIN3/CIN3 lesions similarly into cancer-like and normal-like categories. Distinct methylation changes occur across the genome during the progression of AC and CC with overall similar profiles and add to the evidence suggesting that HPV-driven oncogenesis may result in similar non-random methylomic events. Our findings may lead to identification of potential epigenetic drivers of HPV-associated cancers and also, of potential markers to identify higher risk pre-cancerous lesions.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: Dr. Berglund discloses grants from NIH and from Florida Biomedical Research program, during the conduct of the study. Dr. Eschrich reports grants from the NCI, during the conduct of the study and other relevant financial activities with Cvergenx, Inc., outside the submitted work. Dr. Guha reports grants and personal fees from Johnson & Johnson, grants from Celldex, and other financial activities from Focused Ultrasound Foundation and Varian, outside the submitted work. Dr. Siegel reports grant from the NIH and from the Florida Biomedical Research Program, during the conduct of the study. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures


References
-
- Hariri S, Unger ER, Schafer S, Niccolai LM, Park IU, Bloch KC, et al.. HPV type attribution in high-grade cervical lesions: assessing the potential benefits of vaccines in a population-based evaluation in the United States. Cancer Epidemiol Biomarkers Prev. 2015;24(2):393–9. doi: 10.1158/1055-9965.EPI-14-0649 - DOI - PubMed
-
- Jemal A, Simard EP, Dorell C, Noone AM, Markowitz LE, Kohler B, et al.. Annual Report to the Nation on the Status of Cancer, 1975–2009, featuring the burden and trends in human papillomavirus(HPV)-associated cancers and HPV vaccination coverage levels. Journal of the National Cancer Institute. 2013;105(3):175–201. doi: 10.1093/jnci/djs491 - DOI - PMC - PubMed
-
- McCredie MR, Sharples KJ, Paul C, Baranyai J, Medley G, Jones RW, et al.. Natural history of cervical neoplasia and risk of invasive cancer in women with cervical intraepithelial neoplasia 3: a retrospective cohort study. Lancet Oncol. 2008;9(5):425–34. doi: 10.1016/S1470-2045(08)70103-7 - DOI - PubMed

