The Origin and Contribution of Cancer-Associated Fibroblasts in Colorectal Carcinogenesis
- PMID: 34883119
- PMCID: PMC8881386
- DOI: 10.1053/j.gastro.2021.11.037
The Origin and Contribution of Cancer-Associated Fibroblasts in Colorectal Carcinogenesis
Abstract
Background & aims: Cancer-associated fibroblasts (CAFs) play an important role in colorectal cancer (CRC) progression and predict poor prognosis in CRC patients. However, the cellular origins of CAFs remain unknown, making it challenging to therapeutically target these cells. Here, we aimed to identify the origins and contribution of colorectal CAFs associated with poor prognosis.
Methods: To elucidate CAF origins, we used a colitis-associated CRC mouse model in 5 different fate-mapping mouse lines with 5-bromodeoxyuridine dosing. RNA sequencing of fluorescence-activated cell sorting-purified CRC CAFs was performed to identify a potential therapeutic target in CAFs. To examine the prognostic significance of the stromal target, CRC patient RNA sequencing data and tissue microarray were used. CRC organoids were injected into the colons of knockout mice to assess the mechanism by which the stromal gene contributes to colorectal tumorigenesis.
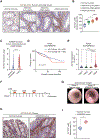
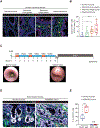
Results: Our lineage-tracing studies revealed that in CRC, many ACTA2+ CAFs emerge through proliferation from intestinal pericryptal leptin receptor (Lepr)+ cells. These Lepr-lineage CAFs, in turn, express melanoma cell adhesion molecule (MCAM), a CRC stroma-specific marker that we identified with the use of RNA sequencing. High MCAM expression induced by transforming growth factor β was inversely associated with patient survival in human CRC. In mice, stromal Mcam knockout attenuated orthotopically injected colorectal tumoroid growth and improved survival through decreased tumor-associated macrophage recruitment. Mechanistically, fibroblast MCAM interacted with interleukin-1 receptor 1 to augment nuclear factor κB-IL34/CCL8 signaling that promotes macrophage chemotaxis.
Conclusions: In colorectal carcinogenesis, pericryptal Lepr-lineage cells proliferate to generate MCAM+ CAFs that shape the tumor-promoting immune microenvironment. Preventing the expansion/differentiation of Lepr-lineage CAFs or inhibiting MCAM activity could be effective therapeutic approaches for CRC.
Keywords: Alpha-Smooth Muscle Actin (αSMA); CD146; Colorectal Cancer; Tumor Microenvironment.
Copyright © 2022 AGA Institute. All rights reserved.
Conflict of interest statement
Disclosures
F.R. is a consultant to or on the advisory board of Agomab, Allergan, AbbVie, Boehringer-Ingelheim, Celgene/BMS, CDISC, Cowen, Genentech, Gilead, Gossamer, Guidepoint, Helmsley, Index Pharma, Jannsen, Koutif, Mestag, Metacrine, Morphic, Origo, Pfizer, Pliant, Prometheus Biosciences, Receptos, RedX, Roche, Samsung, Surrozen, Takeda, Techlab, Theravance, Thetis, and UCB. The remaining authors disclose no conflicts.
Figures




Comment in
-
Origin of CAFs in colorectal cancer.Nat Rev Gastroenterol Hepatol. 2022 Feb;19(2):79. doi: 10.1038/s41575-021-00573-8. Nat Rev Gastroenterol Hepatol. 2022. PMID: 34983959 No abstract available.
References
-
- Kobayashi H, Enomoto A, Woods SL, et al. Cancer-associated fibroblasts in gastrointestinal cancer. Nat Rev Gastroenterol Hepatol 2019;16:282–295. - PubMed
-
- Lee HO, Hong Y, Etlioglu HE, et al. Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nat Genet 2020;52:594–603. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

