Evaluation of multiple analytical methods for SARS-CoV-2 surveillance in wastewater samples
- PMID: 34883175
- PMCID: PMC8648376
- DOI: 10.1016/j.scitotenv.2021.152033
Evaluation of multiple analytical methods for SARS-CoV-2 surveillance in wastewater samples
Abstract
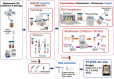
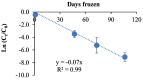
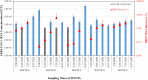
In this study, 14 virus concentration protocols based on centrifugation, filtration, polyethylene glycol (PEG) precipitation and ultrafiltration were tested for their efficacy for the quantification of SARS-CoV-2 in wastewater samples. These protocols were paired with four RNA extraction procedures resulting in a combination of 50 unique approaches. Bovine respiratory syncytial virus (BRSV) was used as a process control and seeded in each wastewater sample subjected to all 50 protocols. The recovery of BRSV obtained through the application of 50 unique approaches ranged from <0.03 to 64.7% (±1.6%). Combination of centrifugation as the solid removal step, ultrafiltration (Amicon-UF-15; 100 kDa cut-off; protocol 9) as the primary virus concentration method, and Zymo Quick-RNA extraction kit provided the highest BRSV recovery (64.7 ± 1.6%). To determine the impact of prolonged storage of large wastewater sample volume (900 mL) at -20 °C on enveloped virus decay, the BRSV seeded wastewaters samples were stored at -20 °C up to 110 days and analyzed using the most efficient concentration (protocol 9) and extraction (Zymo Quick-RNA kit) methods. BRSV RNA followed a first-order decay rate (k = 0.04/h with r2 = 0.99) in wastewater. Finally, 21 wastewater influent samples from five wastewater treatment plants (WWTPs) in southern Maryland, USA were analyzed between May to August 2020 to determine SARS-CoV-2 RNA concentrations. SARS-CoV-2 RNA was quantifiable in 17/21 (81%) of the influent wastewater samples with concentration ranging from 1.10 (±0.10) × 104 to 2.38 (±0.16) × 106 gene copies/L. Among the RT-qPCR assays tested, US CDC N1 assay was the most sensitive followed by US CDC N2, E_Sarbeco, and RdRp assays. Data presented in this study may enhance our understanding on the effective concentration and extraction of SARS-CoV-2 from wastewater.
Keywords: COVID-19; RT-qPCR; SARS-CoV-2; Surveillance; Virus concentration; Wastewater.
Copyright © 2021 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could influence the work and results presented in this paper.
Figures
Similar articles
-
Development of an efficient wastewater testing protocol for high-throughput country-wide SARS-CoV-2 monitoring.Sci Total Environ. 2022 Jun 20;826:154024. doi: 10.1016/j.scitotenv.2022.154024. Epub 2022 Feb 22. Sci Total Environ. 2022. PMID: 35217043 Free PMC article.
-
Evaluation of colorimetric RT-LAMP for screening of SARS-CoV-2 in untreated wastewater.Sci Total Environ. 2024 Jan 10;907:167964. doi: 10.1016/j.scitotenv.2023.167964. Epub 2023 Oct 20. Sci Total Environ. 2024. PMID: 37865239
-
Evaluation of process limit of detection and quantification variation of SARS-CoV-2 RT-qPCR and RT-dPCR assays for wastewater surveillance.Water Res. 2022 Apr 15;213:118132. doi: 10.1016/j.watres.2022.118132. Epub 2022 Feb 3. Water Res. 2022. PMID: 35152136 Free PMC article.
-
A review on detection of SARS-CoV-2 RNA in wastewater in light of the current knowledge of treatment process for removal of viral fragments.J Environ Manage. 2021 Dec 1;299:113563. doi: 10.1016/j.jenvman.2021.113563. Epub 2021 Aug 19. J Environ Manage. 2021. PMID: 34488114 Free PMC article. Review.
-
Approaches applied to detect SARS-CoV-2 in wastewater and perspectives post-COVID-19.J Water Process Eng. 2021 Apr;40:101947. doi: 10.1016/j.jwpe.2021.101947. Epub 2021 Jan 29. J Water Process Eng. 2021. PMID: 35592728 Free PMC article. Review.
Cited by
-
Improved methods for the detection and quantification of SARS-CoV-2 RNA in wastewater.Sci Rep. 2022 May 3;12(1):7201. doi: 10.1038/s41598-022-11187-8. Sci Rep. 2022. PMID: 35504966 Free PMC article.
-
Application of Skimmed-Milk Flocculation Method for Wastewater Surveillance of COVID-19 in Kathmandu, Nepal.Pathogens. 2024 Apr 29;13(5):366. doi: 10.3390/pathogens13050366. Pathogens. 2024. PMID: 38787218 Free PMC article.
-
The potential of long-term wastewater-based surveillance to predict COVID-19 waves peak in Mexico.Water Environ Res. 2025 May;97(5):e70095. doi: 10.1002/wer.70095. Water Environ Res. 2025. PMID: 40396702 Free PMC article.
-
The comparison of decay rates of infectious SARS-CoV-2 and viral RNA in environmental waters and wastewater.Sci Total Environ. 2024 Oct 10;946:174379. doi: 10.1016/j.scitotenv.2024.174379. Epub 2024 Jun 30. Sci Total Environ. 2024. PMID: 38955270
-
Identifying spatiotemporal trends of SARS-CoV-2 RNA in wastewater: from the perspective of upstream and downstream wastewater-based epidemiology (WBE).Environ Sci Pollut Res Int. 2024 Feb;31(8):11576-11590. doi: 10.1007/s11356-023-31769-x. Epub 2024 Jan 14. Environ Sci Pollut Res Int. 2024. PMID: 38221556
References
-
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 - PMC - PubMed
-
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., Gyawali P., Korajkic A., Riddell S., Sherchan S.P., Simpson S.L., Sirikanchana K., Symonds E.M., Verhagen R., Vasan S.S., Kitajima M., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 - PMC - PubMed
-
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Choi P.M., Farkas K., Gyawali P., Hamilton K.A., Haramoto E., Kitajima M., Simpson S.L., Tandukar S., Thomas K., Mueller J.F. Surveillance of SARS-CoV-2 RNA in wastewater: methods optimisation and quality control are crucial for generating reliable public health information. Curr. Opin. Environ. Sci. Health. 2020 doi: 10.1016/j.coesh.2020.09.003. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

