TIN2 is an architectural protein that facilitates TRF2-mediated trans- and cis-interactions on telomeric DNA
- PMID: 34883513
- PMCID: PMC8682769
- DOI: 10.1093/nar/gkab1142
TIN2 is an architectural protein that facilitates TRF2-mediated trans- and cis-interactions on telomeric DNA
Abstract
The telomere specific shelterin complex, which includes TRF1, TRF2, RAP1, TIN2, TPP1 and POT1, prevents spurious recognition of telomeres as double-strand DNA breaks and regulates telomerase and DNA repair activities at telomeres. TIN2 is a key component of the shelterin complex that directly interacts with TRF1, TRF2 and TPP1. In vivo, the large majority of TRF1 and TRF2 are in complex with TIN2 but without TPP1 and POT1. Since knockdown of TIN2 also removes TRF1 and TRF2 from telomeres, previous cell-based assays only provide information on downstream effects after the loss of TRF1/TRF2 and TIN2. Here, we investigated DNA structures promoted by TRF2-TIN2 using single-molecule imaging platforms, including tracking of compaction of long mouse telomeric DNA using fluorescence imaging, atomic force microscopy (AFM) imaging of protein-DNA structures, and monitoring of DNA-DNA and DNA-RNA bridging using the DNA tightrope assay. These techniques enabled us to uncover previously unknown unique activities of TIN2. TIN2S and TIN2L isoforms facilitate TRF2-mediated telomeric DNA compaction (cis-interactions), dsDNA-dsDNA, dsDNA-ssDNA and dsDNA-ssRNA bridging (trans-interactions). Furthermore, TIN2 facilitates TRF2-mediated T-loop formation. We propose a molecular model in which TIN2 functions as an architectural protein to promote TRF2-mediated trans and cis higher-order nucleic acid structures at telomeres.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


 , max and min
, max and min  , 99%, and 1%
, 99%, and 1%  are represented by these symbols in the box plot.
are represented by these symbols in the box plot.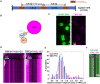

 , max and min
, max and min  , 99% and 1%
, 99% and 1%  are represented by these symbols in the box plot; P < 10–5 *, P < 10–11***.
are represented by these symbols in the box plot; P < 10–5 *, P < 10–11***.


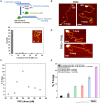

References
-
- Palm W., de Lange T.. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008; 42:301–334. - PubMed
-
- Cech T.R. Beginning to understand the end of the chromosome. Cell. 2004; 116:273–279. - PubMed
-
- Songyang Z., Liu D. Inside the mammalian telomere interactome: regulation and regulatory activities of telomeres. Crit. Rev. Eukaryot. Gene Expr. 2006; 16:103–118. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

