UMPred-FRL: A New Approach for Accurate Prediction of Umami Peptides Using Feature Representation Learning
- PMID: 34884927
- PMCID: PMC8658322
- DOI: 10.3390/ijms222313124
UMPred-FRL: A New Approach for Accurate Prediction of Umami Peptides Using Feature Representation Learning
Abstract
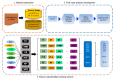
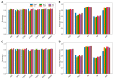
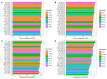
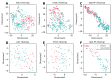
Umami ingredients have been identified as important factors in food seasoning and production. Traditional experimental methods for characterizing peptides exhibiting umami sensory properties (umami peptides) are time-consuming, laborious, and costly. As a result, it is preferable to develop computational tools for the large-scale identification of available sequences in order to identify novel peptides with umami sensory properties. Although a computational tool has been developed for this purpose, its predictive performance is still insufficient. In this study, we use a feature representation learning approach to create a novel machine-learning meta-predictor called UMPred-FRL for improved umami peptide identification. We combined six well-known machine learning algorithms (extremely randomized trees, k-nearest neighbor, logistic regression, partial least squares, random forest, and support vector machine) with seven different feature encodings (amino acid composition, amphiphilic pseudo-amino acid composition, dipeptide composition, composition-transition-distribution, and pseudo-amino acid composition) to develop the final meta-predictor. Extensive experimental results demonstrated that UMPred-FRL was effective and achieved more accurate performance on the benchmark dataset compared to its baseline models, and consistently outperformed the existing method on the independent test dataset. Finally, to aid in the high-throughput identification of umami peptides, the UMPred-FRL web server was established and made freely available online. It is expected that UMPred-FRL will be a powerful tool for the cost-effective large-scale screening of candidate peptides with potential umami sensory properties.
Keywords: bioinformatics; feature representation learning; machine learning; sequence analysis; umami peptide.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
AMYPred-FRL is a novel approach for accurate prediction of amyloid proteins by using feature representation learning.Sci Rep. 2022 May 11;12(1):7697. doi: 10.1038/s41598-022-11897-z. Sci Rep. 2022. PMID: 35546347 Free PMC article.
-
NeuroPred-FRL: an interpretable prediction model for identifying neuropeptide using feature representation learning.Brief Bioinform. 2021 Nov 5;22(6):bbab167. doi: 10.1093/bib/bbab167. Brief Bioinform. 2021. PMID: 33975333
-
Umami-gcForest: Construction of a predictive model for umami peptides based on deep forest.Food Chem. 2025 Feb 1;464(Pt 3):141826. doi: 10.1016/j.foodchem.2024.141826. Epub 2024 Nov 1. Food Chem. 2025. PMID: 39522377
-
Research progress in the screening and evaluation of umami peptides.Compr Rev Food Sci Food Saf. 2022 Mar;21(2):1462-1490. doi: 10.1111/1541-4337.12916. Epub 2022 Feb 24. Compr Rev Food Sci Food Saf. 2022. PMID: 35201672 Review.
-
Comprehensive review and assessment of computational methods for predicting RNA post-transcriptional modification sites from RNA sequences.Brief Bioinform. 2020 Sep 25;21(5):1676-1696. doi: 10.1093/bib/bbz112. Brief Bioinform. 2020. PMID: 31714956 Review.
Cited by
-
Toward a general and interpretable umami taste predictor using a multi-objective machine learning approach.Sci Rep. 2022 Dec 16;12(1):21735. doi: 10.1038/s41598-022-25935-3. Sci Rep. 2022. PMID: 36526644 Free PMC article.
-
Predicting multiple taste sensations with a multiobjective machine learning method.NPJ Sci Food. 2024 Jul 25;8(1):47. doi: 10.1038/s41538-024-00287-6. NPJ Sci Food. 2024. PMID: 39054312 Free PMC article.
-
TROLLOPE: A novel sequence-based stacked approach for the accelerated discovery of linear T-cell epitopes of hepatitis C virus.PLoS One. 2023 Aug 25;18(8):e0290538. doi: 10.1371/journal.pone.0290538. eCollection 2023. PLoS One. 2023. PMID: 37624802 Free PMC article.
-
StackTTCA: a stacking ensemble learning-based framework for accurate and high-throughput identification of tumor T cell antigens.BMC Bioinformatics. 2023 Jul 28;24(1):301. doi: 10.1186/s12859-023-05421-x. BMC Bioinformatics. 2023. PMID: 37507654 Free PMC article.
-
Rapid screening and taste mechanism of novel umami peptides from natural tripeptide database.Food Chem X. 2025 May 23;28:102565. doi: 10.1016/j.fochx.2025.102565. eCollection 2025 May. Food Chem X. 2025. PMID: 40520701 Free PMC article.
References
-
- Dang Y., Gao X., Ma F., Wu X. Comparison of umami taste peptides in water-soluble extractions of Jinhua and Parma hams. LWT-Food Sci. Technol. 2015;60:1179–1186. doi: 10.1016/j.lwt.2014.09.014. - DOI
-
- Wang W., Zhou X., Liu Y. Characterization and evaluation of umami taste: A review. Trends Anal. Chem. 2020;127:115876. doi: 10.1016/j.trac.2020.115876. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

