CYCLON and NPM1 Cooperate within an Oncogenic Network Predictive of R-CHOP Response in DLBCL
- PMID: 34885010
- PMCID: PMC8656558
- DOI: 10.3390/cancers13235900
CYCLON and NPM1 Cooperate within an Oncogenic Network Predictive of R-CHOP Response in DLBCL
Abstract
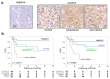
R-CHOP immuno-chemotherapy significantly improved clinical management of diffuse large B-cell lymphoma (DLBCL). However, 30-40% of DLBCL patients still present a refractory disease or relapse. Most of the prognostic markers identified to date fail to accurately stratify high-risk DLBCL patients. We have previously shown that the nuclear protein CYCLON is associated with DLBCL disease progression and resistance to anti-CD20 immunotherapy in preclinical models. We also recently reported that it also represents a potent predictor of refractory disease and relapse in a retrospective DLBCL cohort. However, only sparse data are available to predict the potential biological role of CYCLON and how it might exert its adverse effects on lymphoma cells. Here, we characterized the protein interaction network of CYCLON, connecting this protein to the nucleolus, RNA processing, MYC signaling and cell cycle progression. Among this network, NPM1, a nucleolar multi-functional protein frequently deregulated in cancer, emerged as another potential target related to treatment resistance in DLBCL. Immunohistochemistry evaluation of CYCLON and NPM1 revealed that their co-expression is strongly related to inferior prognosis in DLBCL. More specifically, alternative sub-cellular localizations of the proteins (extra-nucleolar CYCLON and pan-cellular NPM1) represent independent predictive factors specifically associated to R-CHOP refractory DLBCL patients, which could allow them to be orientated towards risk-adapted or novel targeted therapies.
Keywords: CYCLON; NPM1; R-CHOP; R-IPI; nucleolus; prognosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Coiffier B., Thieblemont C., Neste E.V.D., Lepeu G., Plantier I., Castaigne S., Lefort S., Marit G., Macro M., Sebban C., et al. Long-term outcome of patients in the LNH-98.5 trial, the first randomized study comparing rituximab-CHOP to standard CHOP chemotherapy in DLBCL patients: A study by the Groupe d’Etudes des Lymphomes de l’Adulte. Blood. 2010;116:2040–2045. doi: 10.1182/blood-2010-03-276246. - DOI - PMC - PubMed
-
- Swerdlow S.H., Campo E., Pileri S.A., Harris N.L., Stein H., Siebert R., Advani R., Ghielmini M., Salles G.A., Zelenetz A.D., et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–2390. doi: 10.1182/blood-2016-01-643569. - DOI - PMC - PubMed
-
- Rosenwald A., Wright G., Chan W.C., Connors J.M., Campo E., Fisher R.I., Gascoyne R.D., Muller-Hermelink H.K., Smeland E.B., Giltnane J.M., et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N. Engl. J. Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

