Organ Specific Copy Number Variations in Visceral Metastases of Human Melanoma
- PMID: 34885093
- PMCID: PMC8657127
- DOI: 10.3390/cancers13235984
Organ Specific Copy Number Variations in Visceral Metastases of Human Melanoma
Abstract
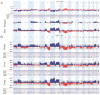
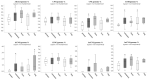
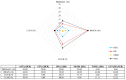
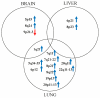
Malignant melanoma is one of the most aggressive skin cancers with high potential of visceral dissemination. Since the information about melanoma genomics is mainly based on primary tumors and lymphatic or skin metastases, an autopsy-based visceral metastasis biobank was established. We used copy number variation arrays (N = 38 samples) to reveal organ specific alterations. Results were partly completed by proteomic analysis. A significant increase of high-copy number gains was found in an organ-specific manner, whereas copy number losses were predominant in brain metastases, including the loss of numerous DNA damage response genes. Amplification of many immune genes was also observed, several of them are novel in melanoma, suggesting that their ectopic expression is possibly underestimated. This "immunogenic mimicry" was exclusive for lung metastasis. We also provided evidence for the possible autocrine activation of c-MET, especially in brain and lung metastases. Furthermore, frequent loss of 9p21 locus in brain metastases may predict higher metastatic potential to this organ. Finally, a significant correlation was observed between BRAF gene copy number and mutant allele frequency, mainly in lung metastases. All of these events may influence therapy efficacy in an organ specific manner, which knowledge may help in alleviating difficulties caused by resistance.
Keywords: BRAF and NRAS mutant allele frequency; DDR deficiency; HGF/MET autocrine activation; distant organ metastasis; immunogenic mimicry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Makohon-Moore A.P., Lipson E.J., Hooper J.E., Zucker A., Hong J., Bielski C.M., Hayashi A., Tokheim C., Baez P., Kappagantula R., et al. The Genetic Evolution of Treatment-Resistant Cutaneous, Acral, and Uveal Melanomas. Clin. Cancer Res. 2021;27:1516–1525. doi: 10.1158/1078-0432.CCR-20-2984. - DOI - PMC - PubMed
-
- Liu L., Bai X., Wang J., Tang X.-R., Wu D.-H., Du S.-S., Du X.-J., Zhang Y.-W., Zhu H.-B., Fang Y., et al. Combination of TMB and CNA Stratifies Prognostic and Predictive Responses to Immunotherapy Across Metastatic Cancer. Clin. Cancer Res. 2019;25:7413–7423. doi: 10.1158/1078-0432.CCR-19-0558. - DOI - PubMed
-
- Reimann J.D.R., Salim S., Velazquez E.F., Wang L., Williams K.M., Flejter W.L., Brooke L., Sunder S., Busam K.J. Comparison of melanoma gene expression score with histopathology, fluorescence in situ hybridization, and SNP array for the classification of melanocytic neoplasms. Mod. Pathol. 2018;31:1733–1743. doi: 10.1038/s41379-018-0087-6. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

