AutoProstate: Towards Automated Reporting of Prostate MRI for Prostate Cancer Assessment Using Deep Learning
- PMID: 34885246
- PMCID: PMC8656605
- DOI: 10.3390/cancers13236138
AutoProstate: Towards Automated Reporting of Prostate MRI for Prostate Cancer Assessment Using Deep Learning
Abstract
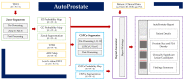
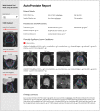
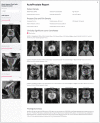
Multiparametric magnetic resonance imaging (mpMRI) of the prostate is used by radiologists to identify, score, and stage abnormalities that may correspond to clinically significant prostate cancer (CSPCa). Automatic assessment of prostate mpMRI using artificial intelligence algorithms may facilitate a reduction in missed cancers and unnecessary biopsies, an increase in inter-observer agreement between radiologists, and an improvement in reporting quality. In this work, we introduce AutoProstate, a deep learning-powered framework for automatic MRI-based prostate cancer assessment. AutoProstate comprises of three modules: Zone-Segmenter, CSPCa-Segmenter, and Report-Generator. Zone-Segmenter segments the prostatic zones on T2-weighted imaging, CSPCa-Segmenter detects and segments CSPCa lesions using biparametric MRI, and Report-Generator generates an automatic web-based report containing four sections: Patient Details, Prostate Size and PSA Density, Clinically Significant Lesion Candidates, and Findings Summary. In our experiment, AutoProstate was trained using the publicly available PROSTATEx dataset, and externally validated using the PICTURE dataset. Moreover, the performance of AutoProstate was compared to the performance of an experienced radiologist who prospectively read PICTURE dataset cases. In comparison to the radiologist, AutoProstate showed statistically significant improvements in prostate volume and prostate-specific antigen density estimation. Furthermore, AutoProstate matched the CSPCa lesion detection sensitivity of the radiologist, which is paramount, but produced more false positive detections.
Keywords: automatic report; computer-aided diagnosis; convolutional neural network; deep learning; lesion classification; lesion detection; magnetic resonance imaging; prostate cancer; segmentation.
Conflict of interest statement
H.U.A. is a paid consultant to Boston Scientific for teaching and training on Rezum for benign prostate hyperplasia treatment and cryotherapy for prostate cancer treatment and is paid for teaching and proctoring HIFU for treating prostate cancer. M.E. receives honoraria from consulting, educational activities, and training from: Sonacare Inc.; NINA Medical; and Angiodynamics Inc. All other authors declare no conflicts of interest.
Figures



Similar articles
-
Factors Influencing Variability in the Performance of Multiparametric Magnetic Resonance Imaging in Detecting Clinically Significant Prostate Cancer: A Systematic Literature Review.Eur Urol Oncol. 2020 Apr;3(2):145-167. doi: 10.1016/j.euo.2020.02.005. Epub 2020 Mar 17. Eur Urol Oncol. 2020. PMID: 32192942 Free PMC article.
-
Prostate cancer detection rate in men undergoing transperineal template-guided saturation and targeted prostate biopsy.Prostate. 2022 Feb;82(3):388-396. doi: 10.1002/pros.24286. Epub 2021 Dec 16. Prostate. 2022. PMID: 34914121 Free PMC article.
-
A Cascaded Deep Learning-Based Artificial Intelligence Algorithm for Automated Lesion Detection and Classification on Biparametric Prostate Magnetic Resonance Imaging.Acad Radiol. 2022 Aug;29(8):1159-1168. doi: 10.1016/j.acra.2021.08.019. Epub 2021 Sep 28. Acad Radiol. 2022. PMID: 34598869 Free PMC article.
-
Added Value of Prostate-specific Antigen Density in Selecting Prostate Biopsy Candidates Among Men with Elevated Prostate-specific Antigen and PI-RADS ≥3 Lesions on Multiparametric Magnetic Resonance Imaging of the Prostate: A Systematic Assessment by PI-RADS Score.Eur Urol Focus. 2024 Jul;10(4):634-640. doi: 10.1016/j.euf.2023.10.006. Epub 2023 Oct 19. Eur Urol Focus. 2024. PMID: 37865591
-
Artificial Intelligence in Magnetic Resonance Imaging-based Prostate Cancer Diagnosis: Where Do We Stand in 2021?Eur Urol Focus. 2022 Mar;8(2):409-417. doi: 10.1016/j.euf.2021.03.020. Epub 2021 Mar 25. Eur Urol Focus. 2022. PMID: 33773964 Review.
Cited by
-
Recent trends in AI applications for pelvic MRI: a comprehensive review.Radiol Med. 2024 Sep;129(9):1275-1287. doi: 10.1007/s11547-024-01861-4. Epub 2024 Aug 3. Radiol Med. 2024. PMID: 39096356 Review.
-
A New Framework for Precise Identification of Prostatic Adenocarcinoma.Sensors (Basel). 2022 Feb 26;22(5):1848. doi: 10.3390/s22051848. Sensors (Basel). 2022. PMID: 35270995 Free PMC article.
-
Anatomically guided self-adapting deep neural network for clinically significant prostate cancer detection on bi-parametric MRI: a multi-center study.Insights Imaging. 2023 Jun 19;14(1):110. doi: 10.1186/s13244-023-01439-0. Insights Imaging. 2023. PMID: 37337101 Free PMC article.
-
Externally validated and clinically useful machine learning algorithms to support patient-related decision-making in oncology: a scoping review.BMC Med Res Methodol. 2025 Feb 21;25(1):45. doi: 10.1186/s12874-025-02463-y. BMC Med Res Methodol. 2025. PMID: 39984835 Free PMC article.
-
Development and clinical utility analysis of a prostate zonal segmentation model on T2-weighted imaging: a multicenter study.Insights Imaging. 2023 Mar 16;14(1):44. doi: 10.1186/s13244-023-01394-w. Insights Imaging. 2023. PMID: 36928683 Free PMC article.
References
-
- Ahmed H.U., El-Shater Bosaily A., Brown L.C., Gabe R., Kaplan R., Parmar M.K., Collaco-Moraes Y., Ward K., Hindley R.G., Freeman A., et al. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): A paired validating confirmatory study. Lancet. 2017;389:815–822. doi: 10.1016/S0140-6736(16)32401-1. - DOI - PubMed
-
- Brembilla G., Dell’Oglio P., Stabile A., Damascelli A., Brunetti L., Ravelli S., Cristel G., Schiani E., Venturini E., Grippaldi D., et al. Interreader variability in prostate MRI reporting using Prostate Imaging Reporting and Data System version 2.1. Eur. Radiol. 2020;30:3383–3392. doi: 10.1007/s00330-019-06654-2. - DOI - PubMed
-
- Stanzione A., Ponsiglione A., Di Fiore G.A., Picchi S.G., Di Stasi M., Verde F., Petretta M., Imbriaco M., Cuocolo R. Prostate Volume Estimation on MRI: Accuracy and Effects of Ellipsoid and Bullet-Shaped Measurements on PSA Density. Acad. Radiol. 2021;28:e219–e226. doi: 10.1016/j.acra.2020.05.014. - DOI - PubMed
-
- Distler F.A., Radtke J.P., Bonekamp D., Kesch C., Schlemmer H.-P., Wieczorek K., Kirchner M., Pahernik S., Hohenfellner M., Hadaschik B.A. The Value of PSA Density in Combination with PI-RADSTM for the Accuracy of Prostate Cancer Prediction. J. Urol. 2017;198:575–582. doi: 10.1016/j.juro.2017.03.130. - DOI - PubMed
-
- Yang X., Lei Y., Wang T., Jiang X., Jani A., Mao H., Curran W., Patel P., Liu T., Wang B. 3D prostate segmentation in MR image using 3D deeply supervised convolutional neural networks. Med. Phys. 2018;45:e582–e583.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

