Effect of human secretory calcium-binding phosphoprotein proline-glutamine rich 1 protein on Porphyromonas gingivalis and identification of its active portions
- PMID: 34887426
- PMCID: PMC8660882
- DOI: 10.1038/s41598-021-02661-w
Effect of human secretory calcium-binding phosphoprotein proline-glutamine rich 1 protein on Porphyromonas gingivalis and identification of its active portions
Abstract
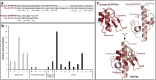
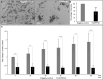
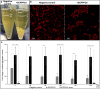
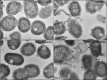
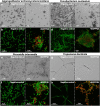
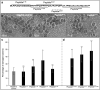
The mouth environment comprises the second most significant microbiome in the body, and its equilibrium is critical in oral health. Secretory calcium-binding phosphoprotein proline-glutamine rich 1 (SCPPPQ1), a protein normally produced by the gingival epithelium to mediate its attachment to teeth, was suggested to be bactericidal. Our aim was to further explore the antibacterial potential of human SCPPPQ1 by characterizing its mode of action and identifying its active portions. In silico analysis showed that it has molecular parallels with antimicrobial peptides. Incubation of Porphyromonas gingivalis, a major periodontopathogen, with the full-length protein resulted in decrease in bacterial number, formation of aggregates and membrane disruptions. Analysis of SCPPPQ1-derived peptides indicated that these effects are sustained by specific regions of the molecule. Altogether, these data suggest that human SCPPPQ1 exhibits antibacterial capacity and provide new insight into its mechanism of action.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Lu M, Xuan S, Wang Z. Oral microbiota: A new view of body health. Food Sci. Hum. Wellness. 2019;8:8–15. doi: 10.1016/j.fshw.2018.12.001. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

