Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing
- PMID: 34887556
- PMCID: PMC9117393
- DOI: 10.1038/s41587-021-01133-w
Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing
Abstract
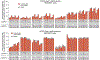
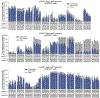
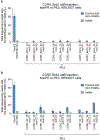
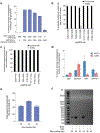
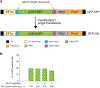
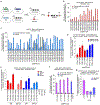
The targeted deletion, replacement, integration or inversion of genomic sequences could be used to study or treat human genetic diseases, but existing methods typically require double-strand DNA breaks (DSBs) that lead to undesired consequences, including uncontrolled indel mixtures and chromosomal abnormalities. Here we describe twin prime editing (twinPE), a DSB-independent method that uses a prime editor protein and two prime editing guide RNAs (pegRNAs) for the programmable replacement or excision of DNA sequences at endogenous human genomic sites. The two pegRNAs template the synthesis of complementary DNA flaps on opposing strands of genomic DNA, which replace the endogenous DNA sequence between the prime-editor-induced nick sites. When combined with a site-specific serine recombinase, twinPE enabled targeted integration of gene-sized DNA plasmids (>5,000 bp) and targeted sequence inversions of 40 kb in human cells. TwinPE expands the capabilities of precision gene editing and might synergize with other tools for the correction or complementation of large or complex human pathogenic alleles.
© 2021. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Declaration of Interests
D.R.L. is a consultant and equity holder of Beam Therapeutics, Prime Medicine, Pairwise Plants, and Chroma Medicine, companies that use genome editing or genome engineering technologies. A.V.A., C.J.P., and J.M.L. are currently employees at Prime Medicine. A.V.A., X.D.G., C.J.P., and D.R. L. have filed patent applications on twinPE and prime editing through the Broad Institute.
Figures








References
Methods References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

