PPIL4 is essential for brain angiogenesis and implicated in intracranial aneurysms in humans
- PMID: 34887573
- PMCID: PMC8768030
- DOI: 10.1038/s41591-021-01572-7
PPIL4 is essential for brain angiogenesis and implicated in intracranial aneurysms in humans
Abstract
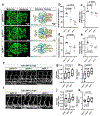
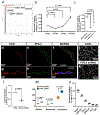
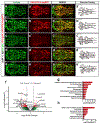
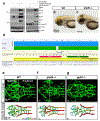
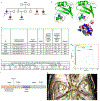
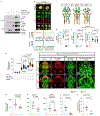
Intracranial aneurysm (IA) rupture leads to subarachnoid hemorrhage, a sudden-onset disease that often causes death or severe disability. Although genome-wide association studies have identified common genetic variants that increase IA risk moderately, the contribution of variants with large effect remains poorly defined. Using whole-exome sequencing, we identified significant enrichment of rare, deleterious mutations in PPIL4, encoding peptidyl-prolyl cis-trans isomerase-like 4, in both familial and index IA cases. Ppil4 depletion in vertebrate models causes intracerebral hemorrhage, defects in cerebrovascular morphology and impaired Wnt signaling. Wild-type, but not IA-mutant, PPIL4 potentiates Wnt signaling by binding JMJD6, a known angiogenesis regulator and Wnt activator. These findings identify a novel PPIL4-dependent Wnt signaling mechanism involved in brain-specific angiogenesis and maintenance of cerebrovascular integrity and implicate PPIL4 gene mutations in the pathogenesis of IA.
© 2021. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures









Similar articles
-
THSD1 (Thrombospondin Type 1 Domain Containing Protein 1) Mutation in the Pathogenesis of Intracranial Aneurysm and Subarachnoid Hemorrhage.Stroke. 2016 Dec;47(12):3005-3013. doi: 10.1161/STROKEAHA.116.014161. Epub 2016 Nov 15. Stroke. 2016. PMID: 27895300 Free PMC article.
-
Genetic study of intracranial aneurysms.Stroke. 2015 Mar;46(3):620-6. doi: 10.1161/STROKEAHA.114.007286. Epub 2015 Feb 3. Stroke. 2015. PMID: 25649796
-
Molecular cloning, structure and expression of a novel nuclear RNA-binding cyclophilin-like gene (PPIL4) from human fetal brain.Cytogenet Cell Genet. 2001;95(1-2):43-7. doi: 10.1159/000057015. Cytogenet Cell Genet. 2001. PMID: 11978968
-
Genetic basis of intracranial aneurysm formation and rupture: clinical implications in the postgenomic era.Neurosurg Focus. 2019 Jul 1;47(1):E10. doi: 10.3171/2019.4.FOCUS19204. Neurosurg Focus. 2019. PMID: 31261114 Review.
-
Intracranial Aneurysms: Pathology, Genetics, and Molecular Mechanisms.Neuromolecular Med. 2019 Dec;21(4):325-343. doi: 10.1007/s12017-019-08537-7. Epub 2019 May 4. Neuromolecular Med. 2019. PMID: 31055715 Free PMC article. Review.
Cited by
-
Kisspeptin-10 Prevents the Development of Cerebral Aneurysms by Reducing the Expression of Egr-1.CNS Neurosci Ther. 2025 May;31(5):e70413. doi: 10.1111/cns.70413. CNS Neurosci Ther. 2025. PMID: 40384564 Free PMC article.
-
Direct blood flow velocity measurements of zebrafish cerebral vessels with a dual-mode light-sheet microscope.Biomed Opt Express. 2025 Jun 2;16(7):2573-2583. doi: 10.1364/BOE.553584. eCollection 2025 Jul 1. Biomed Opt Express. 2025. PMID: 40677814 Free PMC article.
-
The quest to unravel the complex genomics of intracranial aneurysms.Nat Cardiovasc Res. 2022 Apr;1(4):281-282. doi: 10.1038/s44161-022-00051-7. Nat Cardiovasc Res. 2022. PMID: 39196131 No abstract available.
-
A systematic review and meta-analysis of Comaneci/Cascade temporary neck bridging devices for the treatment of intracranial aneurysms.Front Hum Neurosci. 2023 Sep 25;17:1276681. doi: 10.3389/fnhum.2023.1276681. eCollection 2023. Front Hum Neurosci. 2023. PMID: 37817943 Free PMC article.
-
Gene therapy for intracranial aneurysms: systemic review.J Neurointerv Surg. 2025 Jul 14;17(8):859-863. doi: 10.1136/jnis-2024-021843. J Neurointerv Surg. 2025. PMID: 39357890 Review.
References
-
- Vlak MH, Algra A, Brandenburg R & Rinkel GJ Prevalence of unruptured intracranial aneurysms, with emphasis on sex, age, comorbidity, country, and time period: a systematic review and meta-analysis. Lancet Neurol. 10, 626–636 (2011). - PubMed
-
- Lindbohm JV, Kaprio J, Jousilahti P, Salomaa V & Korja M Risk factors of sudden death from subarachnoid hemorrhage. Stroke 48, 2399–2404 (2017). - PubMed
-
- Kissela BM et al. Subarachnoid hemorrhage: a preventable disease with a heritable component. Stroke 33, 1321–1326 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

