A phase 1/2 trial of an immune-modulatory vaccine against IDO/PD-L1 in combination with nivolumab in metastatic melanoma
- PMID: 34887574
- PMCID: PMC8904254
- DOI: 10.1038/s41591-021-01544-x
A phase 1/2 trial of an immune-modulatory vaccine against IDO/PD-L1 in combination with nivolumab in metastatic melanoma
Erratum in
-
Author Correction: A phase 1/2 trial of an immune-modulatory vaccine against IDO/PD-L1 in combination with nivolumab in metastatic melanoma.Nat Med. 2022 Apr;28(4):871. doi: 10.1038/s41591-022-01771-w. Nat Med. 2022. PMID: 35260843 Free PMC article. No abstract available.
Abstract
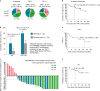
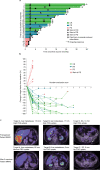
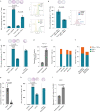
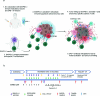
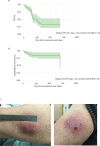
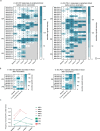
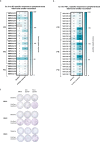
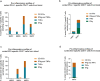
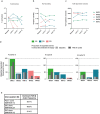
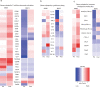
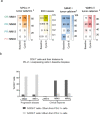
Anti-programmed death (PD)-1 (aPD1) therapy is an effective treatment for metastatic melanoma (MM); however, over 50% of patients progress due to resistance. We tested a first-in-class immune-modulatory vaccine (IO102/IO103) against indoleamine 2,3-dioxygenase (IDO) and PD ligand 1 (PD-L1), targeting immunosuppressive cells and tumor cells expressing IDO and/or PD-L1 (IDO/PD-L1), combined with nivolumab. Thirty aPD1 therapy-naive patients with MM were treated in a phase 1/2 study ( https://clinicaltrials.gov/ , NCT03047928). The primary endpoint was feasibility and safety; the systemic toxicity profile was comparable to that of nivolumab monotherapy. Secondary endpoints were efficacy and immunogenicity; an objective response rate (ORR) of 80% (confidence interval (CI), 62.7-90.5%) was reached, with 43% (CI, 27.4-60.8%) complete responses. After a median follow-up of 22.9 months, the median progression-free survival (PFS) was 26 months (CI, 15.4-69 months). Median overall survival (OS) was not reached. Vaccine-specific responses assessed in vitro were detected in the blood of >93% of patients during vaccination. Vaccine-reactive T cells comprised CD4+ and CD8+ T cells with activity against IDO- and PD-L1-expressing cancer and immune cells. T cell influx of peripherally expanded T cells into tumor sites was observed in responding patients, and general enrichment of IDO- and PD-L1-specific clones after treatment was documented. These clinical efficacy and favorable safety data support further validation in a larger randomized trial to confirm the clinical potential of this immunomodulating approach.
© 2021. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
M.H.A. is named as an inventor on various patent applications relating to therapeutic uses of IDO and PD-L1 peptides. These patent applications are assigned to the company IO Biotech, which is developing immune-modulating cancer treatments. M.H.A. is a founder, shareholder and advisor for IO Biotech. I.M.S. is a cofounder, shareholder and advisor for IO Biotech. I.M.S. has an advisory board relationship with or lectured for Roche, Novartis, MSD, Celgene, Incyte, TILT Bio, Pfizer and BMS AstraZeneca and has received limited grants for translational research from BMS, Roche and Novartis. M.-B.Z. is the CEO, founder and shareholder at IO Biotech. E.M., A.W.P. and E. Ehrnrooth are employees at IO Biotech. E. Ellebaek has received honoraria from BMS, Pierre Fabre, Roche and Kyowa Kirin and travel support from MSD. M.D. has received honoraria from Genzyme, MSD, BMS, Roche and Novartis and travel support from Novartis, MSD, BMS, Roche and Pfizer. The remaining authors declare that they have no conflict of interest.
Figures




References
-
- Robert C, et al. Pembrolizumab versus ipilimumab in advanced melanoma (KEYNOTE-006): post-hoc 5-year results from an open-label, multicentre, randomised, controlled, phase 3 study. Lancet Oncol. 2019;20:1239–1251. - PubMed
-
- Larkin J, et al. Five-year survival with combined nivolumab and ipilimumab in advanced melanoma. N. Engl. J. Med. 2019;381:1535–1546. - PubMed
-
- Ascierto PA, et al. Efficacy of BMS-986016, a monoclonal antibody that targets lymphocyte activation gene-3 (LAG-3), in combination with nivolumab in pts with melanoma who progressed during prior anti-PD-1/PD-L1 therapy (mel prior IO) in all-comer and biomarker-enriched populations. Ann. Oncol. 2017;28:v611–v612.
-
- Angeles L. Warming ‘cold’ melanoma with TLR9 agonists. Cancer Discov. 2018;8:670. - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

