Wnt1 Lineage Specific Deletion of Gpr161 Results in Embryonic Midbrain Malformation and Failure of Craniofacial Skeletal Development
- PMID: 34887903
- PMCID: PMC8650154
- DOI: 10.3389/fgene.2021.761418
Wnt1 Lineage Specific Deletion of Gpr161 Results in Embryonic Midbrain Malformation and Failure of Craniofacial Skeletal Development
Abstract
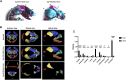
Sonic hedgehog (Shh) signaling regulates multiple morphogenetic processes during embryonic neurogenesis and craniofacial skeletal development. Gpr161 is a known negative regulator of Shh signaling. Nullizygous Gpr161 mice are embryonic lethal, presenting with structural defects involving the neural tube and the craniofacies. However, the lineage specific role of Gpr161 in later embryonic development has not been thoroughly investigated. We studied the Wnt1-Cre lineage specific role of Gpr161 during mouse embryonic development. We observed three major gross morphological phenotypes in Gpr161 cKO (Gpr161 f/f; Wnt1-Cre) fetuses; protrusive tectum defect, encephalocele, and craniofacial skeletal defect. The overall midbrain tissues were expanded and cell proliferation in ventricular zones of midbrain was increased in Gpr161 cKO fetuses, suggesting that protrusive tectal defects in Gpr161 cKO are secondary to the increased proliferation of midbrain neural progenitor cells. Shh signaling activity as well as upstream Wnt signaling activity were increased in midbrain tissues of Gpr161 cKO fetuses. RNA sequencing further suggested that genes in the Shh, Wnt, Fgf and Notch signaling pathways were differentially regulated in the midbrain of Gpr161 cKO fetuses. Finally, we determined that cranial neural crest derived craniofacial bone formation was significantly inhibited in Gpr161 cKO fetuses, which partly explains the development of encephalocele. Our results suggest that Gpr161 plays a distinct role in midbrain development and in the formation of the craniofacial skeleton during mouse embryogenesis.
Keywords: Gpr161; Wnt signaling; craniofacial defects; encephaloceles; midbrain; neural crest cells; sonic hedgehog signaling.
Copyright © 2021 Kim, Robles-Lopez, Cao, Liu, Chothani, Bhavani, Rahman, Mukhopadhyay, Wlodarczyk and Finnell.
Conflict of interest statement
RF formerly held a leadership position in the now defunct TeratOmic Consulting LLC. He also receives travel funds from the journal Reproductive and Developmental Medicine to attend quarterly editorial board meetings. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

