Prolyl endopeptidase-like is a (thio)esterase involved in mitochondrial respiratory chain function
- PMID: 34888501
- PMCID: PMC8634043
- DOI: 10.1016/j.isci.2021.103460
Prolyl endopeptidase-like is a (thio)esterase involved in mitochondrial respiratory chain function
Abstract
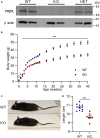
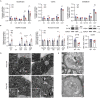
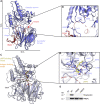
Deficiency of the serine hydrolase prolyl endopeptidase-like (PREPL) causes a recessive metabolic disorder characterized by neonatal hypotonia, feeding difficulties, and growth hormone deficiency. The pathophysiology of PREPL deficiency and the physiological substrates of PREPL remain largely unknown. In this study, we connect PREPL with mitochondrial gene expression and oxidative phosphorylation by analyzing its protein interactors. We demonstrate that the long PREPLL isoform localizes to mitochondria, whereas PREPLS remains cytosolic. Prepl KO mice showed reduced mitochondrial complex activities and disrupted mitochondrial gene expression. Furthermore, mitochondrial ultrastructure was abnormal in a PREPL-deficient patient and Prepl KO mice. In addition, we reveal that PREPL has (thio)esterase activity and inhibition of PREPL by Palmostatin M suggests a depalmitoylating function. We subsequently determined the crystal structure of PREPL, thereby providing insight into the mechanism of action. Taken together, PREPL is a (thio)esterase rather than a peptidase and PREPLL is involved in mitochondrial homeostasis.
Keywords: Molecular biology; Molecular medicine; Structural biology.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Abrami L., Audagnotto M., Ho S., Marcaida M.J., Mesquita F.S., Anwar M.U., Sandoz P.A., Fonti G., Pojer F., Dal Peraro M., et al. Palmitoylated acyl protein thioesterase APT2 deforms membranes to extract substrate acyl chains. Nat. Chem. Biol. 2021;17:438–447. doi: 10.1038/s41589-021-00753-2. - DOI - PMC - PubMed
-
- Bartholdi D., Asadollahi R., Oneda B., Schmitt-Mechelke T., Tonella P., Baumer A., Rauch A. Further delineation of genotype-phenotype correlation in homozygous 2p21 deletion syndromes: first description of patients without cystinuria. Am. J. Med. Genet. 2013;161A:1853–1859. doi: 10.1002/ajmg.a.35994. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

