A Deep-Learning Approach for Inference of Selective Sweeps from the Ancestral Recombination Graph
- PMID: 34888675
- PMCID: PMC8789311
- DOI: 10.1093/molbev/msab332
A Deep-Learning Approach for Inference of Selective Sweeps from the Ancestral Recombination Graph
Abstract
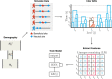
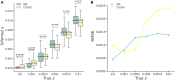
Detecting signals of selection from genomic data is a central problem in population genetics. Coupling the rich information in the ancestral recombination graph (ARG) with a powerful and scalable deep-learning framework, we developed a novel method to detect and quantify positive selection: Selection Inference using the Ancestral recombination graph (SIA). Built on a Long Short-Term Memory (LSTM) architecture, a particular type of a Recurrent Neural Network (RNN), SIA can be trained to explicitly infer a full range of selection coefficients, as well as the allele frequency trajectory and time of selection onset. We benchmarked SIA extensively on simulations under a European human demographic model, and found that it performs as well or better as some of the best available methods, including state-of-the-art machine-learning and ARG-based methods. In addition, we used SIA to estimate selection coefficients at several loci associated with human phenotypes of interest. SIA detected novel signals of selection particular to the European (CEU) population at the MC1R and ABCC11 loci. In addition, it recapitulated signals of selection at the LCT locus and several pigmentation-related genes. Finally, we reanalyzed polymorphism data of a collection of recently radiated southern capuchino seedeater taxa in the genus Sporophila to quantify the strength of selection and improved the power of our previous methods to detect partial soft sweeps. Overall, SIA uses deep learning to leverage the ARG and thereby provides new insight into how selective sweeps shape genomic diversity.
Keywords: ancestral recombination graph; machine learning; positive selection; selective sweep.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures





References
-
- Campagna L, Gronau I, Silveira LF, Siepel A, Lovette IJ.. 2015. Distinguishing noise from signal in patterns of genomic divergence in a highly polymorphic avian radiation. Mol Ecol. 24(16):4238–4251. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

