Omicron variant genome evolution and phylogenetics
- PMID: 34888894
- PMCID: PMC9015349
- DOI: 10.1002/jmv.27515
Omicron variant genome evolution and phylogenetics
Abstract
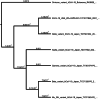
Following the discovery of the SARS-CoV-2 Omicron variant (B.1.1.529), the global COVID-19 outbreak has resurfaced after appearing to be relentlessly spreading over the past 2 years. This new variant showed marked degree of mutation, compared with the previous SARS-CoV-2 variants. This study investigates the evolutionary links between Omicron variant and recently emerged SARS-CoV-2 variants. The entire genome sequences of SARS-CoV-2 variants were obtained, aligned using Clustal Omega, pairwise comparison was computed, differences, identity percent, gaps, and mutations were noted, and the identity matrix was generated. The phylogenetics of Omicron variants were determined using a variety of evolutionary substitution models. The ultrametric and metric clustering methods, such as UPGMA and neighbor-joining (NJ), using nucleotide substitution models that allowed the inclusion of nucleotide transitions and transversions as Kimura 80 models, revealed that the Omicron variant forms a new monophyletic clade that is distant from other SARS-CoV-2 variants. In contrast, the NJ method using a basic nucleotide substitution model such as Jukes-Cantor revealed a close relationship between the Omicron variant and the recently evolved Alpha variant. Based on the percentage of sequence identity, the closest variants were in the following order: Omicron, Alpha, Gamma, Delta, Beta, Mu, and then the SARS-CoV-2 USA isolate. A genome alignment with other variants indicated the greatest number of gaps in the Omicron variant's genome ranging from 43 to 63 gaps. It is possible, given their close relationship to the Alpha variety, that Omicron has been around for much longer than predicted, even though they created a separate monophyletic group. Sequencing initiatives in a systematic and comprehensive manner is highly recommended to study the evolution and mutations of the virus.
Keywords: COVID-19; Omicron variant; SARS-CoV-2; phylogenetics.
© 2021 Wiley Periodicals LLC.
Conflict of interest statement
The authors declare that there are no conflict of interests.
Figures






References
-
- World Health Organization Classification of Omicron (B.1.1.529): SARS‐CoV‐2 Variant of Concern. Accessed November 26, 2021. https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1....
-
- Gao SJ, Guo H, Luo G. Omicron variant (B.1.1.529) of SARS‐CoV‐2, a global urgent public health alert!. J Med Virol. 2021. https://onlinelibrary.wiley.com/doi/10.1002/jmv.27491 - DOI - PMC - PubMed
-
- Kupferschmidt K. Where did ‘weird’ Omicron come from? Science. 2021;374(6572):1179. - PubMed
-
- CLC Genomics Workbench 12.0 (QIAGEN). Accessed December 5, 2021. https://digitalinsights.qiagen.com/
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

