Monoallelic IFT140 pathogenic variants are an important cause of the autosomal dominant polycystic kidney-spectrum phenotype
- PMID: 34890546
- PMCID: PMC8764120
- DOI: 10.1016/j.ajhg.2021.11.016
Monoallelic IFT140 pathogenic variants are an important cause of the autosomal dominant polycystic kidney-spectrum phenotype
Abstract
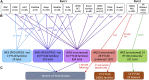
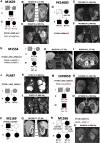
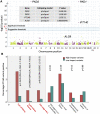
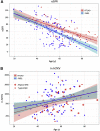
Autosomal dominant polycystic kidney disease (ADPKD), characterized by progressive cyst formation/expansion, results in enlarged kidneys and often end stage kidney disease. ADPKD is genetically heterogeneous; PKD1 and PKD2 are the common loci (∼78% and ∼15% of families) and GANAB, DNAJB11, and ALG9 are minor genes. PKD is a ciliary-associated disease, a ciliopathy, and many syndromic ciliopathies have a PKD phenotype. In a multi-cohort/-site collaboration, we screened ADPKD-diagnosed families that were naive to genetic testing (n = 834) or for whom no PKD1 and PKD2 pathogenic variants had been identified (n = 381) with a PKD targeted next-generation sequencing panel (tNGS; n = 1,186) or whole-exome sequencing (WES; n = 29). We identified monoallelic IFT140 loss-of-function (LoF) variants in 12 multiplex families and 26 singletons (1.9% of naive families). IFT140 is a core component of the intraflagellar transport-complex A, responsible for retrograde ciliary trafficking and ciliary entry of membrane proteins; bi-allelic IFT140 variants cause the syndromic ciliopathy, short-rib thoracic dysplasia (SRTD9). The distinctive monoallelic phenotype is mild PKD with large cysts, limited kidney insufficiency, and few liver cysts. Analyses of the cystic kidney disease probands of Genomics England 100K showed that 2.1% had IFT140 LoF variants. Analysis of the UK Biobank cystic kidney disease group showed probands with IFT140 LoF variants as the third most common group, after PKD1 and PKD2. The proximity of IFT140 to PKD1 (∼0.5 Mb) in 16p13.3 can cause diagnostic confusion, and PKD1 variants could modify the IFT140 phenotype. Importantly, our studies link a ciliary structural protein to the ADPKD spectrum.
Keywords: ADPKD; IFT140; cilia; ciliopathy; intraflagellar transport; monoallelic cystic disease; polycystic kidney disease; short rib thoracic dysplasia.
Copyright © 2021 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.M. reports grants and consulting fees outside the submitted work from Otsuka Pharmaceuticals, Sanofi, Chinook, Goldilocks, Natera, and Palladio. R.D.P. reports clinical trial support from Reata, Kadmon, Sanofi-Genzyme, US Department of Defense; consultant/advisory fees from Otsuka and Sanofi-Genzyme; and is section editor Renal Cystic Disease: UpToDate. J.A.S. has received honorarium from consulting positions from Otsuka Pharmaceuticals, Sanofi, and Takeda. V.E.T. reports grants and/or other fees from Mironid, Blueprint Medicines, Otsuka Pharmaceuticals, Palladio Biosciences, Sanofi Genzyme, Reata, and Regulus Therapeutics, all outside the submitted work.
Figures


References
-
- Cornec-Le Gall E., Alam A., Perrone R.D. Autosomal dominant polycystic kidney disease. Lancet. 2019;393:919–935. - PubMed
-
- Sanchis I.M., Shukoor S., Irazabal M.V., Madsen C.D., Chebib F.T., Hogan M.C., El-Zoghby Z., Harris P.C., Huston J., Brown R.D., Torres V.E. Presymptomatic Screening for Intracranial Aneurysms in Patients with Autosomal Dominant Polycystic Kidney Disease. Clin. J. Am. Soc. Nephrol. 2019;14:1151–1160. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

