CONFIRMS: A Toolkit for Scalable, Black Box Connectome Assessment and Investigation
- PMID: 34891774
- PMCID: PMC9073849
- DOI: 10.1109/EMBC46164.2021.9630109
CONFIRMS: A Toolkit for Scalable, Black Box Connectome Assessment and Investigation
Abstract
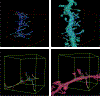
The nanoscale connectomics community has recently generated automated and semi-automated "wiring diagrams" of brain subregions from terabytes and petabytes of dense 3D neuroimagery. This process involves many challenging and imperfect technical steps, including dense 3D image segmentation, anisotropic nonrigid image alignment and coregistration, and pixel classification of each neuron and their individual synaptic connections. As data volumes continue to grow in size, and connectome generation becomes increasingly commonplace, it is important that the scientific community is able to rapidly assess the quality and accuracy of a connectome product to promote dataset analysis and reuse. In this work, we share our scalable toolkit for assessing the quality of a connectome reconstruction via targeted inquiry and large-scale graph analysis, and to provide insights into how such connectome proofreading processes may be improved and optimized in the future. We illustrate the applications and ecosystem on a recent reference dataset.Clinical relevance- Large-scale electron microscopy (EM) data offers a novel opportunity to characterize etiologies and neurological diseases and conditions at an unprecedented scale. EM is useful for low-level analyses such as biopsies; this increased scale offers new possibilities for research into areas such as neural networks if certain bottlenecks and problems are overcome.
Figures




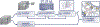
References
-
- Januszewski M, Kornfeld J et al., “High-precision automated reconstruction of neurons with flood-filling networks,” Nature Methods, vol. 15, no. 8, pp. 605–610, Aug. 2018. - PubMed
-
- Buhmann J, Sheridan A et al., “Automatic Detection of Synaptic Partners in a Whole-Brain Drosophila EM Dataset,” bioRxiv, p. 2019.12.12.874172, Mar. 2020.
-
- Funke J, Moreno-Noguer F, Cardona A, and Cook M, “TED: A tolerant edit distance for segmentation evaluation,” ArXiV, Feb. 2016. - PubMed
-
- Schneider-Mizell CM, Bodor AL et al., “Chandelier cell anatomy and function reveal a variably distributed but common signal,” bioRxiv, p. 2020.03.31.018952, Apr. 2020.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
