Long non-coding RNA NORAD/miR-224-3p/MTDH axis contributes to CDDP resistance of esophageal squamous cell carcinoma by promoting nuclear accumulation of β-catenin
- PMID: 34893064
- PMCID: PMC8662861
- DOI: 10.1186/s12943-021-01455-y
Long non-coding RNA NORAD/miR-224-3p/MTDH axis contributes to CDDP resistance of esophageal squamous cell carcinoma by promoting nuclear accumulation of β-catenin
Abstract
Background: Cis-diamminedichloro-platinum (CDDP)-based chemotherapy regimens are the most predominant treatment strategies for patients with esophageal squamous cell carcinoma (ESCC). Dysregulated long non-coding RNAs (lncRNAs) contribute to CDDP resistance, which results in treatment failure in ESCC patients. However, the majority of lncRNAs involved in CDDP resistance in ESCC remain to be elucidated.
Methods: The public Gene Expression Omnibus (GEO) dataset GSE45670 was analysed to reveal potential lncRNAs involved in CDDP resistance of ESCC. Candidate upregulated lncRNAs were detected in ESCC specimens by qRT-PCR to identify crucial lncRNAs. Non-coding RNA activated by DNA damage (NORAD) was selected for further study. Kaplan-Meier analysis and a COX proportional regression model were performed to analyse the potential of NORAD for predicting prognosis of ESCC patients. The role of NORAD in CDDP resistance were determined by conducting gain and loss-of-function experiments in vitro. Fluorescence in situ hybridization (FISH) was performed to determine the subcellular location of NORAD in ESCC cells. A public GEO dataset and bioinformatic algorithms were used to predict the microRNAs (miRNAs) that might be latently sponged by NORAD. qRT-PCR was conducted to verify the expression of candidate miRNAs. Luciferase reporter and Argonaute-2 (Ago2)-RNA immunoprecipitation (RIP) assays were conducted to evaluate the interaction between NORAD and candidate miRNAs. A miRNA rescue experiment was performed to authenticate the NORAD regulatory axis and its effects on CDDP resistance in ESCC cells. Western blotting was conducted to confirm the precise downstream signalling pathway of NORAD. A xenograft mouse model was established to reveal the effect of NORAD on CDDP resistance in vivo.
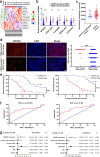
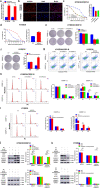
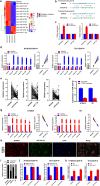
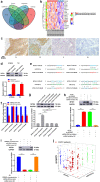
Results: The expression of NORAD was higher in CDDP-resistant ESCC tissues and cells than in CDDP-sensitive tissues and cells. NORAD expression was negatively correlated with the postoperative prognosis of ESCC patients who underwent CDDP-based chemotherapy. NORAD knockdown partially arrested CDDP resistance of ESCC cells. FISH showed that NORAD was located in the cytoplasm in ESCC cells. Furthermore, overlapping results from bioinformatic algorithms analyses and qRT-PCR showed that NORAD could sponge miR-224-3p in ESCC cells. Ago2-RIP demonstrated that NORAD and miR-224-3p occupied the same Ago2 to form an RNA-induced silencing complex (RISC) and subsequently regulated the expression of metadherin (MTDH) in ESCC cells. The NORAD/miR-224-3p/MTDH axis promoted CDDP resistance and progression in ESCC cells by promoting nuclear accumulation of β-catenin in vitro and in vivo.
Conclusions: NORAD upregulates MTDH to promote CDDP resistance and progression in ESCC by sponging miR-224-3p. Our results highlight the potential of NORAD as a therapeutic target in ESCC patients receiving CDDP-based chemotherapy.
Keywords: CDDP resistance; ESCC; MTDH; NORAD; miR-224-3p.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Niksic M, Bonaventure A, Valkov M, Johnson CJ, Esteve J, et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391:1023–1075. - PMC - PubMed
-
- Wang Y, Wang X, Xu G, Gou S. Novel CK2-specific Pt(II) compound reverses Cisplatin-induced resistance by inhibiting Cancer cell Stemness and suppressing DNA damage repair in non-small cell lung Cancer treatments. J Med Chem. 2021;64:4163–4178. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

