Fine-scale population structure and demographic history of British Pakistanis
- PMID: 34893604
- PMCID: PMC8664933
- DOI: 10.1038/s41467-021-27394-2
Fine-scale population structure and demographic history of British Pakistanis
Abstract
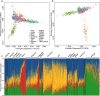
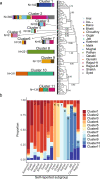
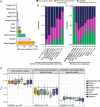
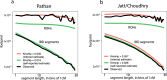
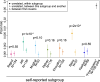
Previous genetic and public health research in the Pakistani population has focused on the role of consanguinity in increasing recessive disease risk, but little is known about its recent population history or the effects of endogamy. Here, we investigate fine-scale population structure, history and consanguinity patterns using genotype chip data from 2,200 British Pakistanis. We reveal strong recent population structure driven by the biraderi social stratification system. We find that all subgroups have had low recent effective population sizes (Ne), with some showing a decrease 15‒20 generations ago that has resulted in extensive identity-by-descent sharing and homozygosity, increasing the risk of recessive disorders. Our results from two orthogonal methods (one using machine learning and the other coalescent-based) suggest that the detailed reporting of parental relatedness for mothers in the cohort under-represents the true levels of consanguinity. These results demonstrate the impact of cultural practices on population structure and genomic diversity in Pakistanis, and have important implications for medical genetic studies.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Bittles, A. H. In Vogel and Motulsky’s Human Genetics (eds Speicher, M. R., Motulsky, A. G. & Antonarakis, S. E.) 507–528 (Springer, 2010).
-
- Office For National Statistics. Census: Ethnic Group, Local Authorities in the United Kingdom (Office For National Statistics, 2011).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

