Type-I collagen produced by distinct fibroblast lineages reveals specific function during embryogenesis and Osteogenesis Imperfecta
- PMID: 34893625
- PMCID: PMC8664945
- DOI: 10.1038/s41467-021-27563-3
Type-I collagen produced by distinct fibroblast lineages reveals specific function during embryogenesis and Osteogenesis Imperfecta
Abstract
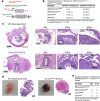
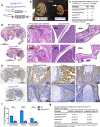
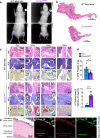
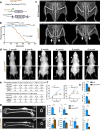
Type I collagen (Col1) is the most abundant protein in mammals. Col1 contributes to 90% of the total organic component of bone matrix. However, the precise cellular origin and functional contribution of Col1 in embryogenesis and bone formation remain unknown. Single-cell RNA-sequencing analysis identifies Fap+ cells and Fsp1+ cells as the major contributors of Col1 in the bone. We generate transgenic mouse models to genetically delete Col1 in various cell lineages. Complete, whole-body Col1 deletion leads to failed gastrulation and early embryonic lethality. Specific Col1 deletion in Fap+ cells causes severe skeletal defects, with hemorrhage, edema, and prenatal lethality. Specific Col1 deletion in Fsp1+ cells results in Osteogenesis Imperfecta-like phenotypes in adult mice, with spontaneous fractures and compromised bone healing. This study demonstrates specific contributions of mesenchymal cell lineages to Col1 production in organogenesis, skeletal development, and bone formation/repair, with potential insights into cell-based therapy for patients with Osteogenesis Imperfecta.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Lohler J, Timpl R, Jaenisch R. Embryonic lethal mutation in mouse collagen I gene causes rupture of blood vessels and is associated with erythropoietic and mesenchymal cell death. Cell. 1984;38:597–607. - PubMed
-
- Zaidi M. Skeletal remodeling in health and disease. Nat. Med. 2007;13:791–801. - PubMed
-
- Marini JC, et al. Osteogenesis imperfecta. Nat. Rev. Dis. Prim. 2017;3:17052. - PubMed
-
- Rauch F, Glorieux FH. Osteogenesis imperfecta. Lancet. 2004;363:1377–1385. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

