Neurocognitive and hypokinetic movement disorder with features of parkinsonism after BCMA-targeting CAR-T cell therapy
- PMID: 34893771
- PMCID: PMC8678323
- DOI: 10.1038/s41591-021-01564-7
Neurocognitive and hypokinetic movement disorder with features of parkinsonism after BCMA-targeting CAR-T cell therapy
Abstract
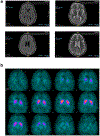
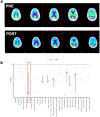
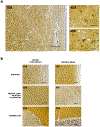
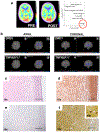
B-cell maturation antigen (BCMA) is a prominent tumor-associated target for chimeric antigen receptor (CAR)-T cell therapy in multiple myeloma (MM). Here, we describe the case of a patient with MM who was enrolled in the CARTITUDE-1 trial ( NCT03548207 ) and who developed a progressive movement disorder with features of parkinsonism approximately 3 months after ciltacabtagene autoleucel BCMA-targeted CAR-T cell infusion, associated with CAR-T cell persistence in the blood and cerebrospinal fluid, and basal ganglia lymphocytic infiltration. We show BCMA expression on neurons and astrocytes in the patient's basal ganglia. Public transcriptomic datasets further confirm BCMA RNA expression in the caudate of normal human brains, suggesting that this might be an on-target effect of anti-BCMA therapy. Given reports of three patients with grade 3 or higher parkinsonism on the phase 2 ciltacabtagene autoleucel trial and of grade 3 parkinsonism in the idecabtagene vicleucel package insert, our findings support close neurological monitoring of patients on BCMA-targeted T cell therapies.
© 2021. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures








References
-
- Kumar SK, et al. Multiple myeloma. Nat Rev Dis Primers 3, 17046 (2017). - PubMed
-
- Madduri D, Dhodapkar MV, Lonial S, Jagannath S & Cho HJ SOHO State of the Art Updates and Next Questions: T-Cell-Directed Immune Therapies for Multiple Myeloma: Chimeric Antigen Receptor-Modified T Cells and Bispecific T-Cell-Engaging Agents. Clin Lymphoma Myeloma Leuk 19, 537–544 (2019). - PubMed
-
- Shah UA & Mailankody S Emerging immunotherapies in multiple myeloma. BMJ 370, m3176 (2020). - PubMed
METHODS-ONLY REFERENCES
-
- Kolde R pheatmap: Pretty Heatmaps. R package version 1.0.12 (2019).
-
- Wickham H ggplot2: Elegant Graphics for Data Analysis, (Springer-Verlag; New York, 2016).
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

