Island Biogeography Revisited: Museomics Reveals Affinities of Shelf Island Birds Determined by Bathymetry and Paleo-Rivers, Not by Distance to Mainland
- PMID: 34893875
- PMCID: PMC8789277
- DOI: 10.1093/molbev/msab340
Island Biogeography Revisited: Museomics Reveals Affinities of Shelf Island Birds Determined by Bathymetry and Paleo-Rivers, Not by Distance to Mainland
Abstract
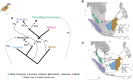
Island biogeography is one of the most powerful subdisciplines of ecology: its mathematical predictions that island size and distance to mainland determine diversity have withstood the test of time. A key question is whether these predictions follow at a population-genomic level. Using rigorous ancient-DNA protocols, we retrieved approximately 1,000 genomic markers from approximately 100 historic specimens of two Southeast Asian songbird complexes from across the Sunda Shelf archipelago collected 1893-1957. We show that the genetic affinities of populations on small shelf islands defy the predictions of geographic distance and appear governed by Earth-historic factors including the position of terrestrial barriers (paleo-rivers) and persistence of corridors (Quaternary land bridges). Our analyses suggest that classic island-biogeographic predictors may not hold well for population-genomic dynamics on the thousands of shelf islands across the globe, which are exposed to dynamic changes in land distribution during Quaternary climate change.
Keywords: Quaternary glacial cycles; Sundaland; ancient DNA; babblers; paleorivers.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures




References
-
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Available from: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
-
- Bird MI, Taylor D, Hunt C.. 2005. Palaeoenvironments of insular Southeast Asia during the Last Glacial Period: a savanna corridor in Sundaland? Quat Sci Rev. 24(20–21):2228–2242.
-
- Borregaard MK, Matthews TJ, Whittaker RJ, Field R.. 2016. The general dynamic model: towards a unified theory of island biogeography? Glob Ecol Biogeogr. 25(7):805–816.

