INTERMEDIUM-C mediates the shade-induced bud growth arrest in barley
- PMID: 34894212
- PMCID: PMC8982414
- DOI: 10.1093/jxb/erab542
INTERMEDIUM-C mediates the shade-induced bud growth arrest in barley
Abstract
Tiller formation is a key agronomic determinant for grain yield in cereal crops. The modulation of this trait is controlled by transcriptional regulators and plant hormones, tightly regulated by external environmental conditions. While endogenous (genetic) and exogenous (environmental factors) triggers for tiller formation have mostly been investigated separately, it has remained elusive how they are integrated into the developmental program of this trait. The transcription factor gene INTERMEDIUM-C (INT-C), which is the barley ortholog of the maize domestication gene TEOSINTE BRANCHED1 (TB1), has a prominent role in regulating tiller bud outgrowth. Here we show that INT-C is expressed in tiller buds, required for bud growth arrest in response to shade. In contrast to wild-type plants, int-c mutant plants are impaired in their shade response and do not stop tiller production after shading. Gene expression levels of INT-C are up-regulated under light-limiting growth conditions, and down-regulated after decapitation. Transcriptome analysis of wild-type and int-c buds under control and shading conditions identified target genes of INT-C that belong to auxin and gibberellin biosynthesis and signaling pathways. Our study identifies INT-C as an integrator of the shade response into tiller formation, which is prerequisite for implementing shading responses in the breeding of cereal crops.
Keywords: INTERMEDIUM-C; Abscisic acid; barley; bud growth arrest; decapitation; shade avoidance; yield.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures

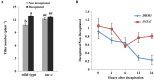




References
-
- Ballaré CL. 1999. Keeping up with the neighbours: phytochrome sensing and other signalling mechanisms. Trends in Plant Science 4, 201. - PubMed
-
- Bustin SA, Benes V, Garson JA, et al. . 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry 55, 611–622. - PubMed
-
- Casal JJ, Sanchez RA, Deregibus VA.. 1986. The effect of plant-density on tillering—the involvement of R/Fr ratio and the proportion of radiation intercepted per plant. Environmental and Experimental Botany 26, 365–371.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

