Interaction between Rag genes results in a unique synergistic transcriptional response that enhances soybean resistance to soybean aphids
- PMID: 34895143
- PMCID: PMC8665634
- DOI: 10.1186/s12864-021-08147-3
Interaction between Rag genes results in a unique synergistic transcriptional response that enhances soybean resistance to soybean aphids
Abstract
Background: Pyramiding different resistance genes into one plant genotype confers enhanced resistance at the phenotypic level, but the molecular mechanisms underlying this effect are not well-understood. In soybean, aphid resistance is conferred by Rag genes. We compared the transcriptional response of four soybean genotypes to aphid feeding to assess how the combination of Rag genes enhanced the soybean resistance to aphid infestation.
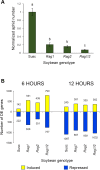
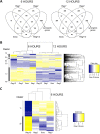
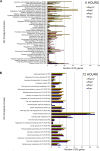
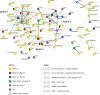
Results: A strong synergistic interaction between Rag1 and Rag2, defined as genes differentially expressed only in the pyramid genotype, was identified. This synergistic effect in the Rag1/2 phenotype was very evident early (6 h after infestation) and involved unique biological processes. However, the response of susceptible and resistant genotypes had a large overlap 12 h after aphid infestation. Transcription factor (TF) analyses identified a network of interacting TF that potentially integrates signaling from Rag1 and Rag2 to produce the unique Rag1/2 response. Pyramiding resulted in rapid induction of phytochemicals production and deposition of lignin to strengthen the secondary cell wall, while repressing photosynthesis. We also identified Glyma.07G063700 as a novel, strong candidate for the Rag1 gene.
Conclusions: The synergistic interaction between Rag1 and Rag2 in the Rag1/2 genotype can explain its enhanced resistance phenotype. Understanding molecular mechanisms that support enhanced resistance in pyramid genotypes could facilitate more directed approaches for crop improvement.
Keywords: Aphid resistance; Gene pyramiding; RNA sequencing; Rag genes; Soybean; Soybean aphids; Synergistic effect.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

