Genome-wide methylation patterns in Marfan syndrome
- PMID: 34895303
- PMCID: PMC8665617
- DOI: 10.1186/s13148-021-01204-4
Genome-wide methylation patterns in Marfan syndrome
Abstract
Background: Marfan syndrome (MFS) is a connective tissue disorder caused by mutations in the Fibrillin-1 gene (FBN1). Here, we undertook the first epigenome-wide association study (EWAS) in patients with MFS aiming at identifying DNA methylation loci associated with MFS phenotypes that may shed light on the disease process.
Methods: The Illumina 450 k DNA-methylation array was used on stored peripheral whole-blood samples of 190 patients with MFS originally included in the COMPARE trial. An unbiased genome-wide approach was used, and methylation of CpG-sites across the entire genome was evaluated. Additionally, we investigated CpG-sites across the FBN1-locus (15q21.1) more closely, since this is the gene defective in MFS. Differentially Methylated Positions (DMPs) and Differentially Methylated Regions (DMRs) were identified through regression analysis. Associations between methylation levels and aortic diameters and presence or absence of 21 clinical features of MFS at baseline were analyzed. Moreover, associations between aortic diameter change, and the occurrence of clinical events (death any cause, type-A or -B dissection/rupture, or aortic surgery) and methylation levels were analyzed.
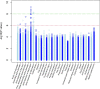
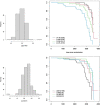
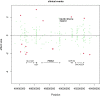
Results: We identified 28 DMPs that are significantly associated with aortic diameters in patients with MFS. Seven of these DMPs (25%) could be allocated to a gene that was previously associated with cardiovascular diseases (HDAC4, IGF2BP3, CASZ1, SDK1, PCDHGA1, DIO3, PTPRN2). Moreover, we identified seven DMPs that were significantly associated with aortic diameter change and five DMP's that associated with clinical events. No significant associations at p < 10-8 or p < 10-6 were found with any of the non-cardiovascular phenotypic MFS features. Investigating DMRs, clusters were seen mostly on X- and Y, and chromosome 18-22. The remaining DMRs indicated involvement of a large family of protocadherins on chromosome 5, which were not reported in MFS before.
Conclusion: This EWAS in patients with MFS has identified a number of methylation loci significantly associated with aortic diameters, aortic dilatation rate and aortic events. Our findings add to the slowly growing literature on the regulation of gene expression in MFS patients.
Keywords: Aortic diameters; Clinical events; EWAS; Marfan syndrome; Methylation loci.
© 2021. The Author(s).
Conflict of interest statement
V. de Waard obtained grants from the AMC Foundation, supporting M.M. van Andel during the conduct of the study. The other authors have nothing to disclose. A.H. Zwinderman and B.J.M. Mulder obtained grants from BBMRI_NL supporting measurements of methylation levels.
Figures
References
-
- Collod-Beroud G, Le Bourdelles S, Ades L, Ala-Kokko L, Booms P, Boxer M, et al. Update of the UMD-FBN1 mutation database and creation of an FBN1 polymorphism database. Hum Mutat. 2003;22(3):199–208. - PubMed
-
- Franken R, Groenink M, de Waard V, Feenstra HM, Scholte AJ, van den Berg MP, et al. Genotype impacts survival in Marfan syndrome. Eur Heart J. 2016;37(43):3285–3290. - PubMed
-
- Franken R, Teixido-Tura G, Brion M, Forteza A, Rodriguez-Palomares J, Gutierrez L, et al. Relationship between fibrillin-1 genotype and severity of cardiovascular involvement in Marfan syndrome. Heart (Br Cardiac Soc) 2017;103(22):1795–1799. - PubMed
-
- Franken R, Heesterbeek TJ, de Waard V, Zwinderman AH, Pals G, Mulder BJM, et al. Diagnosis and genetics of Marfan syndrome. Expert Opin Orphan Drugs. 2014;2(10):1049–1062.
-
- Franken R, den Hartog AW, Radonic T, Micha D, Maugeri A, van Dijk FS, et al. Beneficial outcome of losartan therapy depends on Type of FBN1 mutation in Marfan syndrome. Circ Cardiovasc Genet. 2015;8(2):383–388. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

