Efficient ancestry and mutation simulation with msprime 1.0
- PMID: 34897427
- PMCID: PMC9176297
- DOI: 10.1093/genetics/iyab229
Efficient ancestry and mutation simulation with msprime 1.0
Abstract
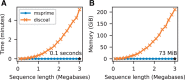
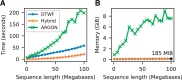
Stochastic simulation is a key tool in population genetics, since the models involved are often analytically intractable and simulation is usually the only way of obtaining ground-truth data to evaluate inferences. Because of this, a large number of specialized simulation programs have been developed, each filling a particular niche, but with largely overlapping functionality and a substantial duplication of effort. Here, we introduce msprime version 1.0, which efficiently implements ancestry and mutation simulations based on the succinct tree sequence data structure and the tskit library. We summarize msprime's many features, and show that its performance is excellent, often many times faster and more memory efficient than specialized alternatives. These high-performance features have been thoroughly tested and validated, and built using a collaborative, open source development model, which reduces duplication of effort and promotes software quality via community engagement.
Keywords: Ancestral Recombination Graphs; coalescent; mutations; simulation.
© The Author(s) 2021. Published by Oxford University Press on behalf of Genetics Society of America.
Figures