Dynamic modulation of enhancer responsiveness by core promoter elements in living Drosophila embryos
- PMID: 34897508
- PMCID: PMC8754644
- DOI: 10.1093/nar/gkab1177
Dynamic modulation of enhancer responsiveness by core promoter elements in living Drosophila embryos
Abstract
Regulatory interactions between enhancers and core promoters are fundamental for the temporal and spatial specificity of gene expression in development. The central role of core promoters is to initiate productive transcription in response to enhancer's activation cues. However, it has not been systematically assessed how individual core promoter elements affect the induction of transcriptional bursting by enhancers. Here, we provide evidence that each core promoter element differentially modulates functional parameters of transcriptional bursting in developing Drosophila embryos. Quantitative live imaging analysis revealed that the timing and the continuity of burst induction are common regulatory steps on which core promoter elements impact. We further show that the upstream TATA also affects the burst amplitude. On the other hand, Inr, MTE and DPE mainly contribute to the regulation of the burst frequency. Genome editing analysis of the pair-rule gene fushi tarazu revealed that the endogenous TATA and DPE are both essential for its correct expression and function during the establishment of body segments in early embryos. We suggest that core promoter elements serve as a key regulatory module in converting enhancer activity into transcription dynamics during animal development.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

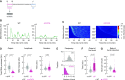




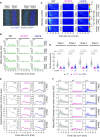
References
-
- Goldberg M.L. Sequence Analysis of Drosophila Histone Genes. 1979; Stanford, CA: Stanford University; Ph.D. Dissertation.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

