Novel Pathogenetic Variants in PTHLH and TRPS1 Genes Causing Syndromic Brachydactyly
- PMID: 34897794
- PMCID: PMC9305952
- DOI: 10.1002/jbmr.4490
Novel Pathogenetic Variants in PTHLH and TRPS1 Genes Causing Syndromic Brachydactyly
Abstract
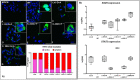
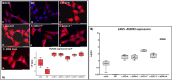
Skeletal disorders, including both isolated and syndromic brachydactyly type E, derive from genetic defects affecting the fine tuning of the network of pathways involved in skeletogenesis and growth-plate development. Alterations of different genes of this network may result in overlapping phenotypes, as exemplified by disorders due to the impairment of the parathyroid hormone/parathyroid hormone-related protein pathway, and obtaining a correct diagnosis is sometimes challenging without a genetic confirmation. Five patients with Albright's hereditary osteodystrophy (AHO)-like skeletal malformations without a clear clinical diagnosis were analyzed by whole-exome sequencing (WES) and novel potentially pathogenic variants in parathyroid hormone like hormone (PTHLH) (BDE with short stature [BDE2]) and TRPS1 (tricho-rhino-phalangeal syndrome [TRPS]) were discovered. The pathogenic impact of these variants was confirmed by in vitro functional studies. This study expands the spectrum of genetic defects associated with BDE2 and TRPS and demonstrates the pathogenicity of TRPS1 missense variants located outside both the nuclear localization signal and the GATA ((A/T)GATA(A/G)-binding zinc-containing domain) and Ikaros-like binding domains. Unfortunately, we could not find distinctive phenotypic features that might have led to an earlier clinical diagnosis, further highlighting the high degree of overlap among skeletal syndromes associated with brachydactyly and AHO-like features, and the need for a close interdisciplinary workout in these rare patients. © 2021 The Authors. Journal of Bone and Mineral Research published by Wiley Periodicals LLC on behalf of American Society for Bone and Mineral Research (ASBMR).
Keywords: CELL/TISSUE SIGNALING - ENDOCRINE PATHWAYS; DISEASES AND DISORDERS OF/RELATED TO BONE; DISORDERS OF CALCIUM/PHOSPHATE METABOLISM; GENETIC RESEARCH; ISORDERS OF CALCIUM/PHOSPHATE METABOLISM; PARATHYROID-RELATED DISORDERS; PTH/VIT D/FGF23.
© 2021 The Authors. Journal of Bone and Mineral Research published by Wiley Periodicals LLC on behalf of American Society for Bone and Mineral Research (ASBMR).
Conflict of interest statement
All the authors declare the absence of any potential conflicts of interest.
Figures