Analysis of Three Architectures for Controlling PTP1B with Light
- PMID: 34898189
- PMCID: PMC8916241
- DOI: 10.1021/acssynbio.1c00398
Analysis of Three Architectures for Controlling PTP1B with Light
Abstract
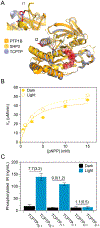
Photosensory domains are powerful tools for placing proteins under optical control, but their integration into light-sensitive chimeras is often challenging. Many designs require structural iterations, and direct comparisons of alternative approaches are rare. This study uses protein tyrosine phosphatase 1B (PTP1B), an influential regulatory enzyme, to compare three architectures for controlling PTPs with light: a protein fusion, an insertion chimera, and a split construct. All three designs permitted optical control of PTP1B activity in vitro (i.e., kinetic assays of purified enzyme) and in mammalian cells; photoresponses measured under both conditions, while different in magnitude, were linearly correlated. The fusion- and insertion-based architectures exhibited the highest dynamic range and maintained native localization patterns in mammalian cells. A single insertion architecture enabled optical control of both PTP1B and TCPTP, but not SHP2, where the analogous chimera was active but not photoswitchable. Findings suggest that PTPs are highly tolerant of domain insertions and support the use of in vitro screens to evaluate different optogenetic designs.
Keywords: light-oxygen-voltage domains; optogenetics; protein engineering; protein tyrosine phosphatases; signaling.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Kim N; Kim JM; Lee M; Kim CY; Chang KY; Heo W. Do. Spatiotemporal control of fibroblast growth factor receptor signals by blue light. Chem. Biol 2014, 21, 903–912. - PubMed
-
- Bantscheff M; Eberhard D; Abraham Y; Bastuck S; Boesche M; Hobson S; Mathieson T; Perrin J; Raida M; Rau C; Reader V; Sweetman G; Bauer A; Bouwmeester T; Hopf C; Kruse U; Neubauer G; Ramsden N; Rick J; Kuster B; Drewes G Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat. Biotechnol 2007, 25, 1035–1044. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

