Transcriptional Profiling of Early and Late Phases of Bovine Tuberculosis
- PMID: 34898250
- PMCID: PMC8853682
- DOI: 10.1128/IAI.00313-21
Transcriptional Profiling of Early and Late Phases of Bovine Tuberculosis
Abstract
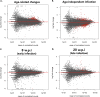
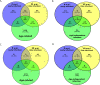
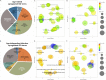
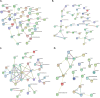
Bovine tuberculosis, caused by Mycobacterium tuberculosis var. bovis (M. bovis), is an important enzootic disease affecting mainly cattle, worldwide. Despite the implementation of national campaigns to eliminate the disease, bovine tuberculosis remains recalcitrant to eradication in several countries. Characterizing the host response to M. bovis infection is crucial for understanding the immunopathogenesis of the disease and for developing better control strategies. To profile the host responses to M. bovis infection, we analyzed the transcriptome of whole blood cells collected from experimentally infected calves with a virulent strain of M. bovis using RNA transcriptome sequencing (RNAseq). Comparative analysis of calf transcriptomes at early (8 weeks) versus late (20 weeks) aerosol infection with M. bovis revealed a divergent and unique profile for each stage of infection. Notably, at the early time point, transcriptional upregulation was observed among several of the top-ranking canonical pathways involved in T-cell chemotaxis. At the late time point, enrichment in the cell mediated cytotoxicity (e.g., Granzyme B) was the predominant host response. These results showed significant change in bovine transcriptional profiles and identified networks of chemokine receptors and monocyte chemoattractant protein (CCL) coregulated genes that underline the host-mycobacterial interactions during progression of bovine tuberculosis in cattle. Further analysis of the transcriptomic profiles identified potential biomarker targets for early and late phases of tuberculosis in cattle. Overall, the identified profiles better characterized identified novel immunomodulatory mechanisms and provided a list of targets for further development of potential diagnostics for tuberculosis in cattle.
Keywords: Mycobacterium tuberculosis var. bovis; RNAseq; bovine tuberculosis; host response; host-pathogen interactions; pathogenesis; transcriptom; transcriptome analysis.
Conflict of interest statement
The authors declare a conflict of interest.
Figures
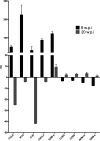
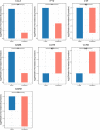
References
-
- Perry BD, Randolph TF, McDermott JJ, Sones KR, Thornton PK. 2002. Investing in animal health research to alleviate poverty. ILRI, Nairobi, Kenya. https://cgspace.cgiar.org/handle/10568/2308. Accessed 25 April 2016.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

