Assessment of Digital Pathology Imaging Biomarkers Associated with Breast Cancer Histologic Grade
- PMID: 34898544
- PMCID: PMC8628688
- DOI: 10.3390/curroncol28060366
Assessment of Digital Pathology Imaging Biomarkers Associated with Breast Cancer Histologic Grade
Abstract
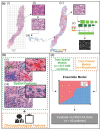
Background: Evaluating histologic grade for breast cancer diagnosis is standard and associated with prognostic outcomes. Current challenges include the time required for manual microscopic evaluation and interobserver variability. This study proposes a computer-aided diagnostic (CAD) pipeline for grading tumors using artificial intelligence.
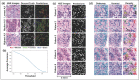
Methods: There were 138 patients included in this retrospective study. Breast core biopsy slides were prepared using standard laboratory techniques, digitized, and pre-processed for analysis. Deep convolutional neural networks (CNNs) were developed to identify the regions of interest containing malignant cells and to segment tumor nuclei. Imaging-based features associated with spatial parameters were extracted from the segmented regions of interest (ROIs). Clinical datasets and pathologic biomarkers (estrogen receptor, progesterone receptor, and human epidermal growth factor 2) were collected from all study subjects. Pathologic, clinical, and imaging-based features were input into machine learning (ML) models to classify histologic grade, and model performances were tested against ground-truth labels at the patient-level. Classification performances were evaluated using receiver-operating characteristic (ROC) analysis.
Results: Multiparametric feature sets, containing both clinical and imaging-based features, demonstrated high classification performance. Using imaging-derived markers alone, the classification performance demonstrated an area under the curve (AUC) of 0.745, while modeling these features with other pathologic biomarkers yielded an AUC of 0.836.
Conclusion: These results demonstrate an association between tumor nuclear spatial features and tumor grade. If further validated, these systems may be implemented into pathology CADs and can assist pathologists to expeditiously grade tumors at the time of diagnosis and to help guide clinical decisions.
Keywords: Nottingham grade; biopsy; breast cancer; computational oncology; imaging biomarkers; tumor.
Conflict of interest statement
K.J.J is a speaker/advisor board/consultant for Amgen, Apo Biologix, Eli Lilly, Esai, Genomic Health, Knight Therapeutics, Pfizer, Roche, Seagen, Merck, Novartis, Purdue Pharma and Viatris. K.J.J received research funding: Eli Lilly, Astra Zeneca. All other authors declare no conflict of interest.
Figures


References
-
- Fitzgibbons P.L., Connolly J.L. Cancer Protocol Templates. [(accessed on 11 August 2021)]. Available online: https://www.cap.org/protocols-and-guidelines/cancer-reporting-tools/canc....
-
- Cortazar P., Zhang L., Untch M., Mehta K., Costantino J.P., Wolmark N., Bonnefoi H., Cameron D., Gianni L., Valagussa P., et al. Pathological complete response and long-term clinical benefit in breast cancer: The CTNeoBC pooled analysis. Lancet. 2014;384:164–172. doi: 10.1016/S0140-6736(13)62422-8. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

