Investigating the role of the carbon storage regulator A (CsrA) in Leptospira spp
- PMID: 34898610
- PMCID: PMC8668096
- DOI: 10.1371/journal.pone.0260981
Investigating the role of the carbon storage regulator A (CsrA) in Leptospira spp
Abstract
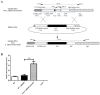
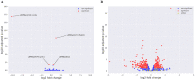
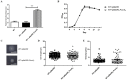
Carbon Storage Regulator A (CsrA) is a well-characterized post-transcriptional global regulator that plays a critical role in response to environmental changes in many bacteria. CsrA has been reported to regulate several metabolic pathways, motility, biofilm formation, and virulence-associated genes. The role of csrA in Leptospira spp., which are able to survive in different environmental niches and infect a wide variety of reservoir hosts, has not been characterized. To investigate the role of csrA as a gene regulator in Leptospira, we generated a L. biflexa csrA deletion mutant (ΔcsrA) and csrA overexpressing Leptospira strains. The ΔcsrA L. biflexa displayed poor growth under starvation conditions. RNA sequencing revealed that in rich medium only a few genes, including the gene encoding the flagellar filament protein FlaB3, were differentially expressed in the ΔcsrA mutant. In contrast, 575 transcripts were differentially expressed when csrA was overexpressed in L. biflexa. Electrophoretic mobility shift assay (EMSA) confirmed the RNA-seq data in the ΔcsrA mutant, showing direct binding of recombinant CsrA to flaB3 mRNA. In the pathogen L. interrogans, we were not able to generate a csrA mutant. We therefore decided to overexpress csrA in L. interrogans. In contrast to the overexpressing strain of L. biflexa, the overexpressing L. interrogans strain had poor motility on soft agar. The overexpressing strain of L. interrogans also showed significant upregulation of the flagellin flaB1, flaB2, and flaB4. The interaction of L. interrogans rCsrA and flaB4 was confirmed by EMSA. Our results demonstrated that CsrA may function as a global regulator in Leptospira spp. under certain conditions that cause csrA overexpression. Interestingly, the mechanisms of action and gene targets of CsrA may be different between non-pathogenic and pathogenic Leptospira strains.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






Similar articles
-
Leptospiral flagellar sheath protein FcpA interacts with FlaA2 and FlaB1 in Leptospira biflexa.PLoS One. 2018 Apr 10;13(4):e0194923. doi: 10.1371/journal.pone.0194923. eCollection 2018. PLoS One. 2018. PMID: 29634754 Free PMC article.
-
The role of CsrA in controls the extracellular electron transfer and biofilm production in Geobacter sulfurreducens.Front Microbiol. 2025 Mar 11;16:1534446. doi: 10.3389/fmicb.2025.1534446. eCollection 2025. Front Microbiol. 2025. PMID: 40135057 Free PMC article.
-
First evidence for gene replacement in Leptospira spp. Inactivation of L. biflexa flaB results in non-motile mutants deficient in endoflagella.Mol Microbiol. 2001 Apr;40(1):189-99. doi: 10.1046/j.1365-2958.2001.02374.x. Mol Microbiol. 2001. PMID: 11298286
-
Comparative and functional genomic analyses of iron transport and regulation in Leptospira spp.J Bacteriol. 2006 Nov;188(22):7893-904. doi: 10.1128/JB.00711-06. Epub 2006 Sep 15. J Bacteriol. 2006. PMID: 16980464 Free PMC article.
-
Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB.Mol Microbiol. 1998 Sep;29(6):1321-30. doi: 10.1046/j.1365-2958.1998.01021.x. Mol Microbiol. 1998. PMID: 9781871 Review.
Cited by
-
Genomic insights into the c-di-GMP signaling and biofilm development in the saprophytic spirochete Leptospira biflexa.Arch Microbiol. 2023 Apr 8;205(5):180. doi: 10.1007/s00203-023-03519-7. Arch Microbiol. 2023. PMID: 37031284
References
-
- Trueba G, Zapata S, Madrid K, Cullen P, Haake D. Cell aggregation: a mechanism of pathogenic Leptospira to survive in fresh water. Int Microbiol. 2004;7(1):35–40. . - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

