The influence of 4-thiouridine labeling on pre-mRNA splicing outcomes
- PMID: 34898625
- PMCID: PMC8668116
- DOI: 10.1371/journal.pone.0257503
The influence of 4-thiouridine labeling on pre-mRNA splicing outcomes
Abstract
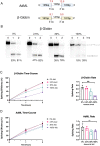
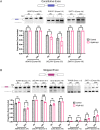
Metabolic labeling is a widely used tool to investigate different aspects of pre-mRNA splicing and RNA turnover. The labeling technology takes advantage of native cellular machineries where a nucleotide analog is readily taken up and incorporated into nascent RNA. One such analog is 4-thiouridine (4sU). Previous studies demonstrated that the uptake of 4sU at elevated concentrations (>50μM) and extended exposure led to inhibition of rRNA synthesis and processing, presumably induced by changes in RNA secondary structure. Thus, it is possible that 4sU incorporation may also interfere with splicing efficiency. To test this hypothesis, we carried out splicing analyses of pre-mRNA substrates with varying levels of 4sU incorporation (0-100%). We demonstrate that increased incorporation of 4sU into pre-mRNAs decreased splicing efficiency. The overall impact of 4sU labeling on pre-mRNA splicing efficiency negatively correlates with the strength of splice site signals such as the 3' and the 5' splice sites. Introns with weaker splice sites are more affected by the presence of 4sU. We also show that transcription by T7 polymerase and pre-mRNA degradation kinetics were impacted at the highest levels of 4sU incorporation. Increased incorporation of 4sU caused elevated levels of abortive transcripts, and fully labeled pre-mRNA is more stable than its uridine-only counterpart. Cell culture experiments show that a small number of alternative splicing events were modestly, but statistically significantly influenced by metabolic labeling with 4sU at concentrations considered to be tolerable (40 μM). We conclude that at high 4sU incorporation rates small, but noticeable changes in pre-mRNA splicing can be detected when splice sites deviate from consensus. Given these potential 4sU artifacts, we suggest that appropriate controls for metabolic labeling experiments need to be included in future labeling experiments.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Dölken L, Ruzsics Z, Rädle B, Friedel CC, Zimmer R, Mages J, et al.. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA [Internet]. 2008. [cited 2021 Apr 1];14(9):1959–72. Available from: http://www.rnajournal.org/cgi/doi/10.1261/rna.1136108. - DOI - PMC - PubMed
-
- Duffy EE, Schofield JA, Simon MD. Gaining insight into transcriptome-wide RNA population dynamics through the chemistry of 4-thiouridine [Internet]. Vol. 10, Wiley Interdisciplinary Reviews: RNA. Blackwell Publishing Ltd; 2019. [cited 2021 Mar 29]. p. e1513. /pmc/articles/PMC6768404/ doi: 10.1002/wrna.1513 - DOI - PMC - PubMed
-
- Herzog VA, Reichholf B, Neumann T, Rescheneder P, Bhat P, Burkard TR, et al.. Thiol-linked alkylation of RNA to assess expression dynamics. Nat Methods [Internet]. 2017. [cited 2021 Mar 22];14(12):1198–204. Available from: http://www.nature.com/authors/editorial_policies/license.html#terms doi: 10.1038/nmeth.4435 - DOI - PMC - PubMed
-
- National Center for Biotechnology Information. PubChem Compound Summary for CID 5359227. Natl Libr Med [Internet]. 2004. [cited 2021 Apr 1];1–49. Available from: https://pubchem.ncbi.nlm.nih.gov/compound/4-Thiouridine
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

