A Core Omnigenic Non-coding Trait Governing Dex-Induced Osteoporotic Effects Identified Without DEXA
- PMID: 34899306
- PMCID: PMC8651565
- DOI: 10.3389/fphar.2021.750959
A Core Omnigenic Non-coding Trait Governing Dex-Induced Osteoporotic Effects Identified Without DEXA
Abstract
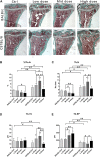
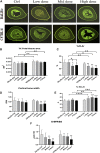
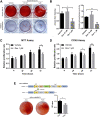
Iatrogenic glucocorticoid (GC)-induced osteoporosis (GIO) is an idiosyncratic form of secondary osteoporosis. Genetic predisposition among individuals may give rise to variant degree of phenotypic changes but there has yet been a documented unified pathway to explain the idiosyncrasy. In this study, we argue that the susceptibility to epigenetic changes governing molecular cross talks along the BMP and PI3K/Akt pathway may underline how genetic background dictate GC-induced bone loss. Concordantly, osteoblasts from BALB/c or C57BL/6 neonatal mice were treated with dexamethasone for transcriptome profiling. Furthermore, we also confirmed that GC-pre-conditioned mesenchymal stem cells (MSCs) would give rise to defective osteogenesis by instigating epigenetic changes which affected the accessibility of enhancer marks. In line with these epigenetic changes, we propose that GC modulates a key regulatory network involving the scavenger receptor Cd36 in osteoblasts pre-conditioning pharmacological idiosyncrasy in GIO.
Keywords: bone loss; bone morphogenetic protein signaling pathway; cis-expression quantitative trait loci; glucocorticoid; glucocorticoid-induced osteoporosis; osteoblasts differentiation.
Copyright © 2021 Lu, Cai, Luo, Wang, Fung, Jia, Yu, Chan, Miu and Xiao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Dexamethasone suppresses osteogenesis of osteoblast via the PI3K/Akt signaling pathway in vitro and in vivo.J Recept Signal Transduct Res. 2019 Feb;39(1):80-86. doi: 10.1080/10799893.2019.1625061. Epub 2019 Jun 18. J Recept Signal Transduct Res. 2019. PMID: 31210570
-
Gastrodin protects MC3T3-E1 osteoblasts from dexamethasone-induced cellular dysfunction and promotes bone formation via induction of the NRF2 signaling pathway.Int J Mol Med. 2018 Apr;41(4):2059-2069. doi: 10.3892/ijmm.2018.3414. Epub 2018 Jan 23. Int J Mol Med. 2018. PMID: 29393365 Free PMC article.
-
Dexamethasone shifts bone marrow stromal cells from osteoblasts to adipocytes by C/EBPalpha promoter methylation.Cell Death Dis. 2013 Oct 3;4(10):e832. doi: 10.1038/cddis.2013.348. Cell Death Dis. 2013. PMID: 24091675 Free PMC article.
-
Glucocorticoid-Induced Osteoporosis.Adv Exp Med Biol. 2015;872:179-215. doi: 10.1007/978-1-4939-2895-8_8. Adv Exp Med Biol. 2015. PMID: 26215995 Free PMC article. Review.
-
The shift in the balance between osteoblastogenesis and adipogenesis of mesenchymal stem cells mediated by glucocorticoid receptor.Stem Cell Res Ther. 2019 Dec 5;10(1):377. doi: 10.1186/s13287-019-1498-0. Stem Cell Res Ther. 2019. PMID: 31805987 Free PMC article. Review.
Cited by
-
CUL1 exacerbates glucocorticoid-induced osteoporosis by enhancing ASAP1 ubiquitination.Hormones (Athens). 2025 Mar;24(1):259-274. doi: 10.1007/s42000-024-00599-y. Epub 2024 Sep 17. Hormones (Athens). 2025. PMID: 39287759
References
-
- Björkegren J. L. M., Kovacic J. C., Dudley J. T., Schadt E. E. (2015). Genome-Wide Significant Loci: How Important Are They? Systems Genetics to Understand Heritability of Coronary Artery Disease and Other Common Complex Disorders. J. Am. Coll. Cardiol. 65 (8), 830–845. 10.1016/j.jacc.2014.12.033 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

