The Evolution of Regulatory Elements in the Emerging Promoter-Variant Strains of HIV-1 Subtype C
- PMID: 34899661
- PMCID: PMC8660095
- DOI: 10.3389/fmicb.2021.779472
The Evolution of Regulatory Elements in the Emerging Promoter-Variant Strains of HIV-1 Subtype C
Abstract
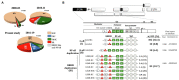
In a multicentric, observational, investigator-blinded, and longitudinal clinical study of 764 ART-naïve subjects, we identified nine different promoter variant strains of HIV-1 subtype C (HIV-1C) emerging in the Indian population, with some of these variants being reported for the first time. Unlike several previous studies, our work here focuses on the evolving viral regulatory elements, not the coding sequences. The emerging viral strains contain additional copies of the existing transcription factor binding sites (TFBS), including TCF-1α/LEF-1, RBEIII, AP-1, and NF-κB, created by sequence duplication. The additional TFBS are genetically diverse and may blur the distinction between the modulatory region of the promoter and the viral enhancer. In a follow-up analysis, we found trends, but no significant associations between any specific variant promoter and prognostic markers, probably because the emerging viral strains might not have established mono infections yet. Illumina sequencing of four clinical samples containing a coinfection indicated the domination of one strain over the other and establishing a stable ratio with the second strain at the follow-up time points. Since a single promoter regulates viral gene expression and constitutes the master regulatory circuit with Tat, the acquisition of additional and variant copies of the TFBS may significantly impact viral latency and latent reservoir characteristics. Further studies are urgently warranted to understand how the diverse TFBS profiles of the viral promoter may modulate the characteristics of the latent reservoir, especially following the initiation of antiretroviral therapy.
Keywords: HIV-1; evolution; latency; sequence duplication; subtype C.
Copyright © 2021 Bhange, Prasad, Singh, Prajapati, Maurya, Gopalan, Nadig, Chaturbhuj, Jayaseelan, Dinesha, Ahamed, Singh, Brahmaiah, Mehta, Gohil, Balakrishnan, Das, Dias, Gangakhedkar, Mehendale, Paranjape, Saravanan, Shet, Solomon, Thakar and Ranga.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Bachu M., Mukthey A. B., Murali R. V., Cheedarla N., Mahadevan A., Shankar S. K., et al. . (2012b). Sequence insertions in the HIV type 1 subtype C viral promoter predominantly generate an additional NF-κB binding site. AIDS Res. Hum. Retrovir. 28, 1362–1368. doi: 10.1089/AID.2011.0388, PMID: - DOI - PMC - PubMed
-
- Bachu M., Yalla S., Asokan M., Verma A., Neogi U., Sharma S., et al. . (2012a). Multiple NF-κB sites in HIV-1 subtype C long terminal repeat confer superior magnitude of transcription and thereby the enhanced viral predominance. J. Biol. Chem. 287, 44714–44735. doi: 10.1074/jbc.M112.397158, PMID: - DOI - PMC - PubMed
-
- Berry I. M., Ribeiro R., Kothari M., Athreya G., Daniels M., Lee H. Y., et al. . (2007). Unequal evolutionary rates in the human immunodeficiency virus type 1 (HIV-1) pandemic: the evolutionary rate of HIV-1 slows down when the epidemic rate increases. J. Virol. 81, 10625–10635. doi: 10.1128/JVI.00985-07, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

