Helicobacter pylori virulence factors: relationship between genetic variability and phylogeographic origin
- PMID: 34900406
- PMCID: PMC8628625
- DOI: 10.7717/peerj.12272
Helicobacter pylori virulence factors: relationship between genetic variability and phylogeographic origin
Abstract
Background: Helicobacter pylori is a pathogenic bacteria that colonize the gastrointestinal tract from human stomachs and causes diseases including gastritis, peptic ulcers, gastric lymphoma (MALT), and gastric cancer, with a higher prevalence in developing countries. Its high genetic diversity among strains is caused by a high mutation rate, observing virulence factors (VFs) variations in different geographic lineages. This study aimed to postulate the genetic variability associated with virulence factors present in the Helicobacter pylori strains, to identify the relationship of these genes with their phylogeographic origin.
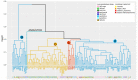
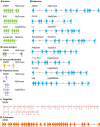
Methods: The complete genomes of 135 strains available in NCBI, from different population origins, were analyzed using bioinformatics tools, identifying a high rate; as well as reorganization events in 87 virulence factor genes, divided into seven functional groups, to determine changes in position, number of copies, nucleotide identity and size, contrasting them with their geographical lineage and pathogenic phenotype.
Results: Bioinformatics analyses show a high rate of gene annotation errors in VF. Analysis of genetic variability of VFs shown that there is not a direct relationship between the reorganization and geographic lineage. However, regarding the pathogenic phenotype demonstrated in the analysis of many copies, size, and similarity when dividing the strains that possess and not the cag pathogenicity island (cagPAI), having a higher risk of developing gastritis and peptic ulcer was evidenced. Our data has shown that the analysis of the overall genetic variability of all VFs present in each strain of H. pylori is key information in understanding its pathogenic behavior.
Keywords: Comparative genomics; Helicobacter pylori; Pathogenicity; Phylogeography; Virulence factors.
© 2021 Rodriguez et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Evolution of Helicobacter spp: variability of virulence factors and their relationship to pathogenicity.PeerJ. 2022 Aug 29;10:e13120. doi: 10.7717/peerj.13120. eCollection 2022. PeerJ. 2022. PMID: 36061745 Free PMC article.
-
Disease-specific Helicobacter pylori virulence factors: the unfulfilled promise.Helicobacter. 2000;5 Suppl 1:S3-9; discussion S27-31. doi: 10.1046/j.1523-5378.2000.0050s1003.x. Helicobacter. 2000. PMID: 10828748 Review.
-
HELICOBACTER PYLORI cagA VIRULENCE GENE AND SEVERE ESOGASTRODUODENAL DISEASES: IS THERE AN ASSOCIATION?Arq Gastroenterol. 2021 Oct-Dec;58(4):468-475. doi: 10.1590/S0004-2803.202100000-85. Arq Gastroenterol. 2021. PMID: 34909852
-
Geographic differences and the role of cagA gene in gastroduodenal diseases associated with Helicobacter pylori infection.Rev Esp Enferm Dig. 2001 Jul;93(7):471-80. Rev Esp Enferm Dig. 2001. PMID: 11685943 Review. English, Spanish.
-
Genomes of Helicobacter pylori from native Peruvians suggest admixture of ancestral and modern lineages and reveal a western type cag-pathogenicity island.BMC Genomics. 2006 Jul 27;7:191. doi: 10.1186/1471-2164-7-191. BMC Genomics. 2006. PMID: 16872520 Free PMC article.
Cited by
-
Identification, Genome Sequencing, and Characterizations of Helicobacter pylori Sourced from Pakistan.Microorganisms. 2023 Oct 29;11(11):2658. doi: 10.3390/microorganisms11112658. Microorganisms. 2023. PMID: 38004670 Free PMC article.
-
Antibiotic Resistance and Genetic Determinants of Helicobacter pylori in Oman: Insights from Phenotypic and Whole-Genome Analysis.Int J Mol Sci. 2025 Jun 12;26(12):5628. doi: 10.3390/ijms26125628. Int J Mol Sci. 2025. PMID: 40565090 Free PMC article.
References
-
- Abu-Taleb AMF, Abdelattef RS, Abdel-Hady AA, Omran FH, El-Korashi LA, Abdel-Aziz El-Hady H, El-Gebaly AM. Prevalence of Helicobacter pylori cagA and iceA genes and their association with gastrointestinal diseases. International Journal of Microbiology. 2018;2018(4):4809093–4809097. doi: 10.1155/2018/4809093. - DOI - PMC - PubMed
-
- Alam J, Sarkar A, Karmakar BC, Ganguly M, Paul S, Mukhopadhyay AK. Novel virulence factor dupA of Helicobacter pylori as an important risk determinant for disease manifestation: an overview. World Journal of Gastroenterology. 2020;26(32):4739–4752. doi: 10.3748/wjg.v26.i32.4739. - DOI - PMC - PubMed
-
- Allan E, Dorrell N, Foynes S, Anyim M, Wren BW. Mutational analysis of genes encoding the early flagellar components of Helicobacter pylori: evidence for transcriptional regulation of flagellin a biosynthesis. Journal of Bacteriology. 2000;182(18):5274–5277. doi: 10.1128/JB.182.18.5274-5277.2000. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

