Prime Editing for Inherited Retinal Diseases
- PMID: 34901928
- PMCID: PMC8656220
- DOI: 10.3389/fgeed.2021.775330
Prime Editing for Inherited Retinal Diseases
Abstract
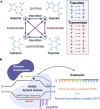
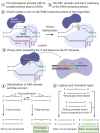
Inherited retinal diseases (IRDs) are chronic, hereditary disorders that lead to progressive degeneration of the retina. Disease etiology originates from a genetic mutation-inherited or de novo-with a majority of IRDs resulting from point mutations. Given the plethora of IRDs, to date, mutations that cause these dystrophies have been found in approximately 280 genes. However, there is currently only one FDA-approved gene augmentation therapy, Luxturna (voretigene neparvovec-rzyl), available to patients with RPE65-mediated retinitis pigmentosa (RP). Although clinical trials for other genes are underway, these techniques typically involve gene augmentation rather than genome surgery. While gene augmentation therapy delivers a healthy copy of DNA to the cells of the retina, genome surgery uses clustered regularly interspaced short palindromic repeats (CRISPR)-based technology to correct a specific genetic mutation within the endogenous genome sequence. A new technique known as prime editing (PE) applies a CRISPR-based technology that possesses the potential to correct all twelve possible transition and transversion mutations as well as small insertions and deletions. EDIT-101, a CRISPR-based therapy that is currently in clinical trials, uses double-strand breaks and nonhomologous end joining to remove the IVS26 mutation in the CEP290 gene. Preferably, PE does not cause double-strand breaks nor does it require any donor DNA repair template, highlighting its unparalleled efficiency. Instead, PE uses reverse transcriptase and Cas9 nickase to repair mutations in the genome. While this technique is still developing, with several challenges yet to be addressed, it offers promising implications for the future of IRD treatment.
Keywords: CRISPR/Cas9 systems; Ophthalmology; adeno-associated viral (AAV) vectors; gene editing; inherited retinal diseases (IRD); prime editing; retinal degeneration.
Copyright © 2021 Costa, Levi, Eulau, Tsai and Quinn.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Aida T., Wilde J. J., Yang L., Hou Y., Li M., Xu D., et al. (2020). Prime Editing Primarily Induces Undesired Outcomes in Mice. bioRxiv. 10.1101/2020.08.06.239723 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

