RdRp inhibitors and COVID-19: Is molnupiravir a good option?
- PMID: 34902743
- PMCID: PMC8654603
- DOI: 10.1016/j.biopha.2021.112517
RdRp inhibitors and COVID-19: Is molnupiravir a good option?
Abstract
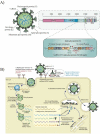
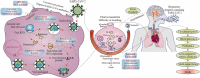
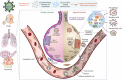
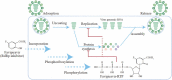
Rapid changes in the viral genome allow viruses to evade threats posed by the host immune response or antiviral drugs, and can lead to viral persistence in the host cells. RNA-dependent RNA polymerase (RdRp) is an essential enzyme in RNA viruses, which is involved in RNA synthesis through the formation of phosphodiester bonds. Therefore, in RNA viral infections such as SARS-CoV-2, RdRp could be a crucial therapeutic target. The present review discusses the promising application of RdRp inhibitors, previously approved or currently being tested in human clinical trials, in the treatment of RNA virus infections. Nucleoside inhibitors (NIs) bind to the active site of RdRp, while nonnucleoside inhibitors (NNIs) bind to allosteric sites. Given the absence of highly effective drugs for the treatment of COVID-19, the discovery of an efficient treatment for this pandemic is an urgent concern for researchers around the world. We review the evidence for molnupiravir (MK-4482, EIDD-2801), an antiviral drug originally designed for Alphavirus infections, as a potential preventive and therapeutic agent for the management of COVID-19. At the beginning of this pandemic, molnupiravir was in preclinical development for seasonal influenza. When COVID-19 spread dramatically, the timeline for development was accelerated to focus on the treatment of this pandemic. Real time consultation with regulators took place to expedite this program. We summarize the therapeutic potential of RdRp inhibitors, and highlight molnupiravir as a new small molecule drug for COVID-19 treatment.
Keywords: Clinical trials; RdRp inhibitors, Molnupiravir; SARS-CoV-2; Small molecule drug.
Copyright © 2021 The Authors. Published by Elsevier Masson SAS.. All rights reserved.
Conflict of interest statement
Authors declare no conflicts of interest.
Figures
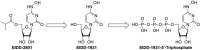
Similar articles
-
Discovery, Development, and Patent Trends on Molnupiravir: A Prospective Oral Treatment for COVID-19.Molecules. 2021 Sep 24;26(19):5795. doi: 10.3390/molecules26195795. Molecules. 2021. PMID: 34641339 Free PMC article. Review.
-
Safety of RNA-Dependent RNA Polymerase Inhibitors, Molnupiravir and VV116, for Oral Treatment of COVID-19: A Meta-Analysis.Iran J Med Sci. 2024 May 1;49(5):275-285. doi: 10.30476/IJMS.2024.99837.3196. eCollection 2024 May. Iran J Med Sci. 2024. PMID: 38751873 Free PMC article. Review.
-
Developing a direct acting, orally available antiviral agent in a pandemic: the evolution of molnupiravir as a potential treatment for COVID-19.Curr Opin Virol. 2021 Oct;50:17-22. doi: 10.1016/j.coviro.2021.06.003. Epub 2021 Jun 18. Curr Opin Virol. 2021. PMID: 34271264 Free PMC article. Review.
-
Molnupiravir: A new candidate for COVID-19 treatment.Pharmacol Res Perspect. 2022 Feb;10(1):e00909. doi: 10.1002/prp2.909. Pharmacol Res Perspect. 2022. PMID: 34968008 Free PMC article. Review.
-
Molnupiravir for the treatment of COVID-19.Drugs Today (Barc). 2022 Jul;58(7):335-350. doi: 10.1358/dot.2022.58.7.3419558. Drugs Today (Barc). 2022. PMID: 35851869
Cited by
-
Medicinal chemistry strategies towards the development of non-covalent SARS-CoV-2 Mpro inhibitors.Acta Pharm Sin B. 2024 Jan;14(1):87-109. doi: 10.1016/j.apsb.2023.08.004. Epub 2023 Aug 9. Acta Pharm Sin B. 2024. PMID: 38239241 Free PMC article. Review.
-
The Role of Cytokines and Chemokines in Severe Acute Respiratory Syndrome Coronavirus 2 Infections.Front Immunol. 2022 Apr 7;13:832394. doi: 10.3389/fimmu.2022.832394. eCollection 2022. Front Immunol. 2022. PMID: 35464491 Free PMC article. Review.
-
The impact of SARS-CoV-2 3CL protease mutations on nirmatrelvir inhibitory efficiency. Computational insights into potential resistance mechanisms.Chem Sci. 2023 Feb 14;14(10):2686-2697. doi: 10.1039/d2sc06584c. eCollection 2023 Mar 8. Chem Sci. 2023. PMID: 36908962 Free PMC article.
-
Potential Anti-SARS-CoV-2 Prodrugs Activated by Phosphorylation and Their Role in the Aged Population.Molecules. 2023 Mar 2;28(5):2332. doi: 10.3390/molecules28052332. Molecules. 2023. PMID: 36903575 Free PMC article. Review.
-
Rapid review and meta-analysis of adverse events associated with molnupiravir in patients with COVID-19.Br J Clin Pharmacol. 2022 Oct;88(10):4403-4411. doi: 10.1111/bcp.15449. Epub 2022 Jul 11. Br J Clin Pharmacol. 2022. PMID: 35762036 Free PMC article. Review.
References
-
- Paules C.I., Marston H.D., Fauci A.S. Coronavirus infections-more than just the common cold. JAMA. 2020;323(8):707–708. - PubMed
-
- Marston H.D., Folkers G.K., Morens D.M., Fauci A.S. Emerging viral diseases: confronting threats with new technologies. Sci. Transl. Med. 2014;6(253) 253ps10. - PubMed
-
- Brown A.J., Won J.J., Graham R.L., Dinnon K.H., 3rd, Sims A.C., Feng J.Y., Cihlar T., Denison M.R., Baric R.S., Sheahan T.P. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antivir. Res. 2019;169 - PMC - PubMed
-
- Menachery V.D., Yount BL Jr, Jr., Debbink K., Agnihothram S., Gralinski L.E., Plante J.A., Graham R.L., Scobey T., Ge X.Y., Donaldson E.F., Randell S.H., Lanzavecchia A., Marasco W.A., Shi Z.L., Baric R.S. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015;21(12):1508–1513. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

