Diverse tumorigenic consequences of human papillomavirus integration in primary oropharyngeal cancers
- PMID: 34903527
- PMCID: PMC8744672
- DOI: 10.1101/gr.275911.121
Diverse tumorigenic consequences of human papillomavirus integration in primary oropharyngeal cancers
Abstract
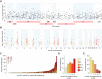
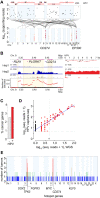
Human papillomavirus (HPV) causes 5% of all cancers and frequently integrates into host chromosomes. The HPV oncoproteins E6 and E7 are necessary but insufficient for cancer formation, indicating that additional secondary genetic events are required. Here, we investigate potential oncogenic impacts of virus integration. Analysis of 105 HPV-positive oropharyngeal cancers by whole-genome sequencing detects virus integration in 77%, revealing five statistically significant sites of recurrent integration near genes that regulate epithelial stem cell maintenance (i.e., SOX2, TP63, FGFR, MYC) and immune evasion (i.e., CD274). Genomic copy number hyperamplification is enriched 16-fold near HPV integrants, and the extent of focal host genomic instability increases with their local density. The frequency of genes expressed at extreme outlier levels is increased 86-fold within ±150 kb of integrants. Across 95% of tumors with integration, host gene transcription is disrupted via intragenic integrants, chimeric transcription, outlier expression, gene breaking, and/or de novo expression of noncoding or imprinted genes. We conclude that virus integration can contribute to carcinogenesis in a large majority of HPV-positive oropharyngeal cancers by inducing extensive disruption of host genome structure and gene expression.
© 2022 Symer et al.; Published by Cold Spring Harbor Laboratory Press.
Figures





References
-
- Bernard E, Pons-Salort M, Favre M, Heard I, Delarocque-Astagneau E, Guillemot D, Thiébaut AC. 2013. Comparing human papillomavirus prevalences in women with normal cytology or invasive cervical cancer to rank genotypes according to their oncogenic potential: a meta-analysis of observational studies. BMC Infect Dis 13: 373. 10.1186/1471-2334-13-373 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
