Genomic and transcriptomic characterization of heterogeneous immune subgroups of microsatellite instability-high colorectal cancers
- PMID: 34903553
- PMCID: PMC8672019
- DOI: 10.1136/jitc-2021-003414
Genomic and transcriptomic characterization of heterogeneous immune subgroups of microsatellite instability-high colorectal cancers
Abstract
Background: Colorectal cancers (CRCs) with microsatellite instability-high (MSI-H) are hypermutated tumors and are generally regarded as immunogenic. However, their heterogeneous immune responses and underlying molecular characteristics remain largely unexplained.
Methods: We conducted a retrospective analysis of 73 primary MSI-H CRC tissues to characterize heterogeneous immune subgroups. Based on combined tumor-infiltrating lymphocyte (TIL) immunoscore and tertiary lymphoid structure (TLS) activity, MSI-H CRCs were classified into immune-high, immune-intermediate, and immune-low subgroups. Of these, the immune-high and immune-low subgroups were further analyzed using whole-exome and transcriptome sequencing.
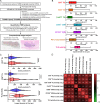
Results: We found considerable variations in immune parameters between MSI-H CRCs, and immune subgrouping of MSI-H CRCs was performed accordingly. The TIL densities and TLS activities of immune-low MSI-H CRCs were comparable to those of an immune-low or immune-intermediate subgroup of microsatellite-stable CRCs. There were remarkable differences between immune-high and immune-low MSI-H CRCs, including their pathological features (medullary vs mucinous), genomic alterations (tyrosine kinase fusions vs KRAS mutations), and activated signaling pathways (immune-related vs Wnt and Notch signaling), whereas no significant differences were found in tumor mutational burden (TMB) and neoantigen load. The immune-low MSI-H CRCs were subdivided by the consensus molecular subtype (CMS1 vs CMS3) with different gene expression signatures (mesenchymal/stem-like vs epithelial/goblet-like), suggesting distinct immune evasion mechanisms. Angiogenesis and CD200 were identified as potential therapeutic targets in immune-low CMS1 and CMS3 MSI-H CRCs, respectively.
Conclusions: MSI-H CRCs are immunologically heterogeneous, regardless of TMB. The unusual immune-low MSI-H CRCs are characterized by mucinous histology, KRAS mutations, and Wnt/Notch activation, and can be further divided into distinct gene expression subtypes, including CMS4-like CMS1 and CMS3. Our data provide novel insights into precise immunotherapeutic strategies for subtypes of MSI-H tumors.
Keywords: gastrointestinal neoplasms; gene expression profiling; immunohistochemistry; lymphocytes; tumor microenvironment; tumor-infiltrating.
© Author(s) (or their employer(s)) 2021. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures



References
-
- Xiao Y, Freeman GJ. The microsatellite instable subset of colorectal cancer is a particularly good candidate for checkpoint blockade immunotherapy. Cancer Discov 2015;5:16–18. 10.1158/2159-8290.CD-14-1397 - DOI - PMC - PubMed
-
- Llosa NJ, Cruise M, Tam A, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov 2015;5:43–51. 10.1158/2159-8290.CD-14-0863 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
