High-impact rare genetic variants in severe schizophrenia
- PMID: 34903660
- PMCID: PMC8713775
- DOI: 10.1073/pnas.2112560118
High-impact rare genetic variants in severe schizophrenia
Abstract
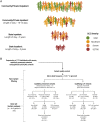
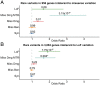
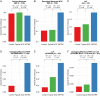
Extreme phenotype sequencing has led to the identification of high-impact rare genetic variants for many complex disorders but has not been applied to studies of severe schizophrenia. We sequenced 112 individuals with severe, extremely treatment-resistant schizophrenia, 218 individuals with typical schizophrenia, and 4,929 controls. We compared the burden of rare, damaging missense and loss-of-function variants between severe, extremely treatment-resistant schizophrenia, typical schizophrenia, and controls across mutation intolerant genes. Individuals with severe, extremely treatment-resistant schizophrenia had a high burden of rare loss-of-function (odds ratio, 1.91; 95% CI, 1.39 to 2.63; P = 7.8 × 10-5) and damaging missense variants in intolerant genes (odds ratio, 2.90; 95% CI, 2.02 to 4.15; P = 3.2 × 10-9). A total of 48.2% of individuals with severe, extremely treatment-resistant schizophrenia carried at least one rare, damaging missense or loss-of-function variant in intolerant genes compared to 29.8% of typical schizophrenia individuals (odds ratio, 2.18; 95% CI, 1.33 to 3.60; P = 1.6 × 10-3) and 25.4% of controls (odds ratio, 2.74; 95% CI, 1.85 to 4.06; P = 2.9 × 10-7). Restricting to genes previously associated with schizophrenia risk strengthened the enrichment with 8.9% of individuals with severe, extremely treatment-resistant schizophrenia carrying a damaging missense or loss-of-function variant compared to 2.3% of typical schizophrenia (odds ratio, 5.48; 95% CI, 1.52 to 19.74; P = 0.02) and 1.6% of controls (odds ratio, 5.82; 95% CI, 3.00 to 11.28; P = 2.6 × 10-8). These results demonstrate the power of extreme phenotype case selection in psychiatric genetics and an approach to augment schizophrenia gene discovery efforts.
Keywords: genomics; rare variants; schizophrenia; treatment-resistant schizophrenia.
Conflict of interest statement
Competing interest statement: D.B.G. is a founder of and holds equity in Q State Biosciences and Praxis Therapeutics; holds equity in Apostle Inc., and serves as a consultant to AstraZeneca, Gilead Sciences, GoldFinch Bio and Gossamer Bio. J.A.L. does not accept any personal financial remuneration for consulting, speaking, or research activities from any pharmaceutical, biotechnology, or medical device companies. He receives funding and medication supplies for investigator-initiated research from Denovo, Taisho, and Cerevel, and company sponsored phase II, III, and IV studies from Alkermes, Sunovion, and Boehringer Ingelheim, which does not contribute to his compensation. He is a consultant or advisory board member of Intracellular Therapies, Takeda, Karuna, Pear Therapeutics, Systems-1, and Psychogenics for which he receives no remuneration. He is a paid consultant for Signant Health, a clinical research technology and services organization, and holds a patent from Repligen that yields no royalties. R.S.D. is a paid consultant for AstraZeneca. X.W. is a cofounder and stock owner at Waypoint Bio. The other authors declare no competing interests.
Figures

References
-
- GBD 2015 Disease and Injury Incidence and Prevalence Collaborators, Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990-2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet 388, 1545–1602 (2016). - PMC - PubMed
-
- Sullivan P. F., Kendler K. S., Neale M. C., Schizophrenia as a complex trait: Evidence from a meta-analysis of twin studies. Arch. Gen. Psychiatry 60, 1187–1192 (2003). - PubMed
-
- Singh T., et al. , Exome sequencing identifies rare coding variants in 10 genes which confer substantial risk for schizophrenia. medRxiv [Preprint] (2020). 10.1101/2020.09.18.20192815. Accessed 14 October 2020. - DOI
Publication types
MeSH terms
Grants and funding
- T32 MH018870/MH/NIMH NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- R01 MH049487/MH/NIMH NIH HHS/United States
- K23 MH121669/MH/NIMH NIH HHS/United States
- R01 MH086873/MH/NIMH NIH HHS/United States
- U01 MH105653/MH/NIMH NIH HHS/United States
- U01 MH105670/MH/NIMH NIH HHS/United States
- R01 MH085548/MH/NIMH NIH HHS/United States
- R01 AG051346/AG/NIA NIH HHS/United States
- TL1 TR001875/TR/NCATS NIH HHS/United States
- R01 MH071523/MH/NIMH NIH HHS/United States
- U01 MH105641/MH/NIMH NIH HHS/United States
- U01 MH105573/MH/NIMH NIH HHS/United States
- R01 MH031340/MH/NIMH NIH HHS/United States
- R01 MH085542/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

